Tutorial 12: Signal Space Projections (SSP)
Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet
As previously said, the frequency filters are not adapted to remove artifacts that are transient or overlapping in frequency domain with the brain signals of interest. Other approaches exist to correct for those artifacts, based on the spatial signature of the artifacts.
If an event is very reproducible and occurs always at the same position (eg. eye blinks and heartbeats), the sensors will always record the same values when it occurs. We can identify the topographies corresponding to this artifact (ie. the spatial distributions of values at one time point) and remove them from the recordings. This spatial decomposition is the basic idea behind two widely used approaches: the SSP (Signal-Space Projection) and ICA (Independent Component Analysis) methods.
This introduction tutorial will focus on the SSP approach, as it is a lot simpler and faster but still very efficient for removing blinks and heartbeats from MEG and high-density EEG recordings. The interface for running ICA decompositions is very similar and will be described in an advanced tutorial.
Contents
Overview
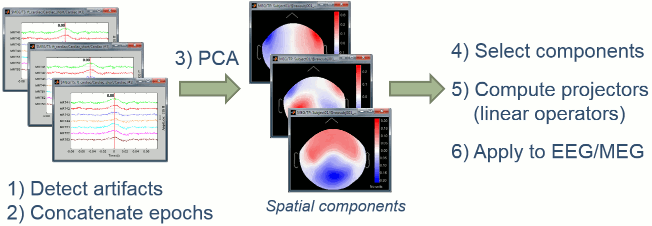
The general SSP objective is to identify the sensor topragraphies that are typical of a specific artifact, then to create spatial projectors to remove the contributions of these topographies from the recordings.
- We start by identifying many examples of the artifact we are trying to remove. This is what we've been doing in the previous tutorial with the creation of the "cardiac" and "blink" events.
- We extract a short time window around each of these event markers and concatenate in time all those small blocks of recordings.
- We run a principle components analysis (PCA) on the concatenated artifacts in order to get a decomposition in various spatial components (number of components = number of sensors).
- If it works well, we can find in the first few principal components some topographies that are very specific of the type of artifact we are targetting. We select those components to remove.
- We compute a linear projector for each spatial component to remove and save them in the database (in the "Link to raw file"). They are not applied immediately to the recordings.
Whenever some recordings are read from this file, the SSP projectors are applied on the fly to remove the artifact contributions. This approach is fast and memory efficient.
The order matters
This procedure has to be repeated separately for each artifact type. The order in which you process the artifacts matters, because for removing the second artifact we typically use the recordings cleaned with the first set of SSP projectors. We have to decide which one to process first.
It works best if each artifact is defined precisely and as independently as possible from the other artifacts. If the two artifacts happen simulateneously, the SSP projectors calculated for the blink may contain some of the heartbeat topography and vice versa. When trying to remove the second artifact, we might not be able to isolate it clearly anymore.
Because the heart beats every second or so, there is a high chance that when the subject blinks there is a heartbeat not too far away in the recordings. Therefore a significant number of the blinks will be contaminated with heartbeats. But we have usually a lot of "clean" heartbeats, we can start by removing those ones. To isolate correctly these two common artifacts, we recommend the following procedure:
- Remove the markers "cardiac" that are occurring during a blink (done in the previous tutorial),
Compute the cardiac SSP (with no eye movements, because we removed the co-occurring events),
Compute the blink SSP (with no heartbeats, because they've already been taken care of).
If you have multiple modalities recorded simultaneously, for example MEG and EEG, you should run twice this entire procedure, once for the EEG only and once for the MEG only. You will always get better results if you process the different types of sensors separately. Same thing when processing Elekta-Neuromag recordings: process separately the magnetometers (MEG MAG) and the gradiometers (MEG GRAD).
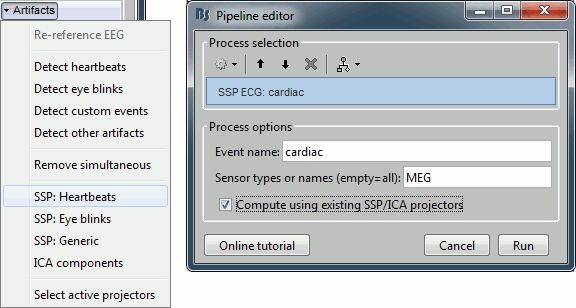
SSP: Heartbeats
Double-click on the link to show the MEG sensors for Run #01.
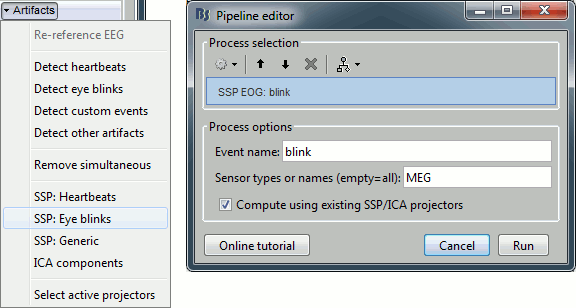
In the Record tab, select the menu: "Artifacts > SSP: Heartbeats".
Event name: Name of the event to use to calculate the projectors, enter "cardiac"
Sensor types: Type of sensors for which the projection should be calculated ("MEG"). Note that you will always get better results if you process the different types of sensors separately.
Compute using existing SSP projectors: You have the option to calculate the projectors from the raw recordings, or from the recordings filtered with the previously computed SSP projectors. Unless you have a good reason for not considering the existing projectors, you should select this option.
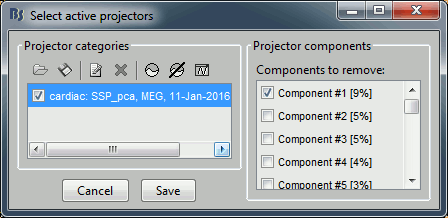
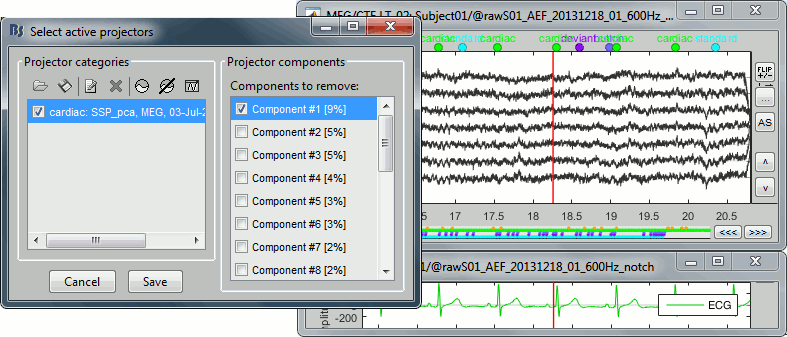
After the computation is done, a new figure is displayed, that lets you select the active projectors.
On the left: The projector categories where each row represents the result of an execution of this process (usually one for each sensor type and each artifact).
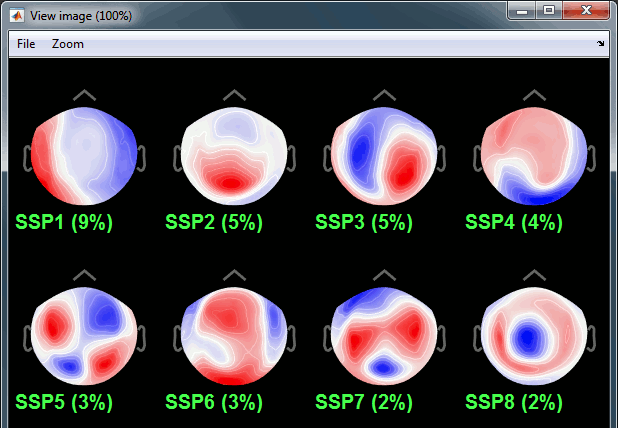
On the right: The spatial components returned by the PCA decomposition. The percentage indicated between brackets is the singular value for this each component, normalized for this decomposition (percentage = Si / sum(Si), see technical details at the end of this page).
Percentage: More practically, it indicates the amount of signal that was captured by the component during the decomposition. The highest it is, the more the component is representative of the artifact recordings that were used to calculate it. In the good cases, you would typically see one to three components with values that are significantly higher that the others.
- When a component is selected, it means that it is removed from the recordings. A spatial projector is computed and applied to the recordings on the fly when reading from the continuous file.
Default selection: The software selects the components with values superior to 12% and leaves the others unselected. This threshold is completely empirical and depends on the acquisition device, you should always review manually the components that you want to remove.
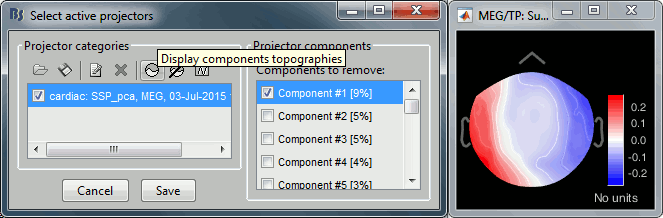
Evaluate the components
For the cardiac event, none of the components show a value superior to 12%, therefore the entire "cardiac" projector category was unselected. This doesn't mean that none of those components is actually interesting for us. The percentage indicated for the first value (9%) is much higher than the following ones (5%, 5%, 4%, 3%...), this could indicate that it targets relatively well the cardiac artifact. Let's investigate this in more details.
- Select the category ("cardiac") to enable the list on the right.
Click on the first component, then click on the toolbar button [Display component topography]. This menu shows the spatial distribution of the values recorded by the MEG sensors for this component. Note that you don't have to select the component (ie. check the box) to display it. This topography seems to correspond to a strong dipolar activity located relatively far from the sensor array, it matches the type of artifact we expect from the heart activity.
The second button "Display component topography [No magnetic interpolation]" produces the same figure but without the reinterpolation of the magnetic fields that is typically applied to the MEG recordings in Brainstorm, it may help understand some difficult cases. This magnetic interpolation will be detailed later in the introduction tutorials.
You can display multiple components in the same figure: select them at the same time in list (holding the Shift/Ctrl/Cmd button of your keyboard) and then click on the button "Display topography".
No other strong components looks like it could be related with the heartbeats.
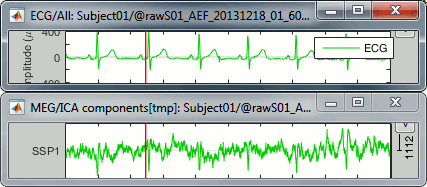
The last button in the toolbar [Display component time series], opens a figure that represents the evolution of the contribution of this component over time. The higher the value, the more present the selected topography in the recordings. Click on it to show the component #1, and display the ECG signal at the same time (right-click on the file > ECG > Display time series).
We observe that the "SSP1" trace correlates relatively well with the ECG trace, we captured most the ECG peaks with this component.
However, the component seems also to capture much more signal than just the heartbeats: many alpha oscillations and some of the ocular activity. The example below shows a blink in the EOG, ECG and SSP component #1.
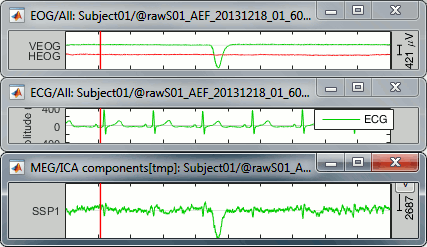
If you remove this component from the recordings, you can expect to see most of the artifacts related with the cardiac activity to go away, but you will also remove additional signal elements that were not really well identified. The job is done but it causes some unwanted side effects.
- It is in general possible to refine the SSP decomposition by going back to the selection of "cardiac" markers that we used to compute it. You could look at all the ECG peaks individually and remove the markers located in segments of recordings that are noisier or that contain a lot of alpha activity (~10Hz). You would need to delete this SSP decomposition and run again the same process.
- Alternatively, or if you don't manage to extract a clean cardiac component with a PCA/SSP decomposition, you could try to run an ICA decomposition instead. Note that for some subjects, the cardiac artifact is not very strong and could be simply ignored in the analysis.
Evaluate the correction
The topography of the component #1 looks like it represents a cardiac topography and its temporal evolution shows peaks where we identified heartbeats. It is therefore a good candidate for removal, we just need to make sure the signals look good after the correction before validating this choice.
- Show the left-temporal MEG sensors (CTF LT) and select/unselect the first SSP component.
- Repeat this for different time windows, to make sure that the cardiac peaks in the MEG sensors really diseappear when the projector #1 is selected and that the rest is not altered too much.
No correction:
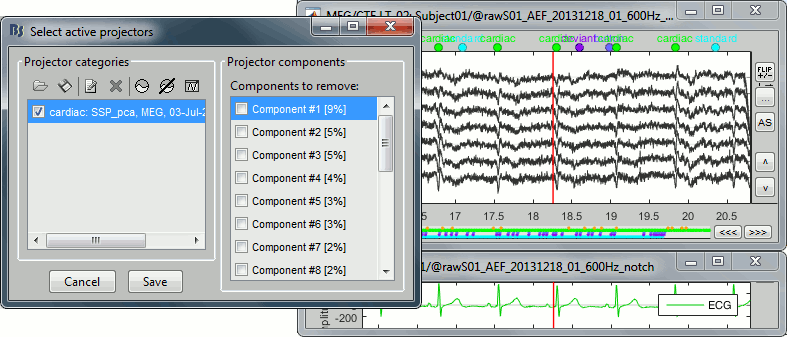
Cardiac component #1 removed:
In this example we will consider that the current decomposition is good enough.
Make sure you select the component #1, then click on [Save] to validate the modifications.After this window is closed, you can always open it again from the Record tab with the menu Artifacts > Select active projectors. At this stage of the analysis, you can modify the list of projectors applied to the recordings at any time.
SSP: Eye blinks
Let's try the same thing with the eye blinks.
Select the process "SSP > Compute SSP: Eye blinks"
Run it on the event type "blink", that indicates the peaks of the EOG signal.
You see now a new category of projectors. It is selected by default because its first component shows a percentage higher than 12% (empirical and somewhat random threshold). Based on the distribution of values, this first component is most likely a good representation of the artifact we are trying to remove. The second one could be a good candidate as well.
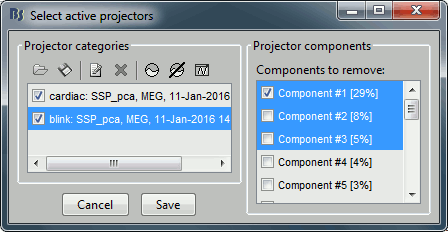
Select the first three components and display their topographies:
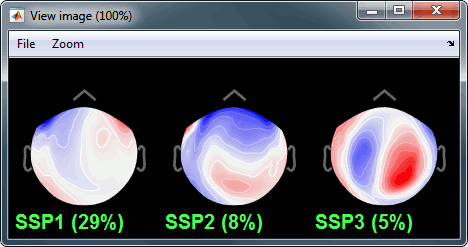
Component #1: Most likely a blink,
Component #2: Probably a saccade (other type of eye movement we were not targetting),
- Component #3: Not related with the eye movements (maybe related with the alpha activity).
Display the times series for those three components, together with the EOG signals. You have to uncheck temporarily the component #1 to be able to display its signal. When it is checked, it is removed from the signal threfore it is corresponds to a flat trace.
The figure below shows the EOG and SSP values between 318s and 324s. The SSP1 trace matches the blink observed in VEOG and SSP2 matches the saccade observed in HEOG.
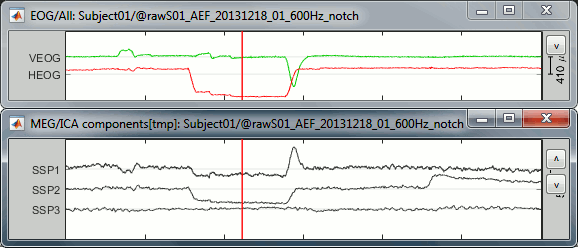
Observe the activity when there is no component selected:
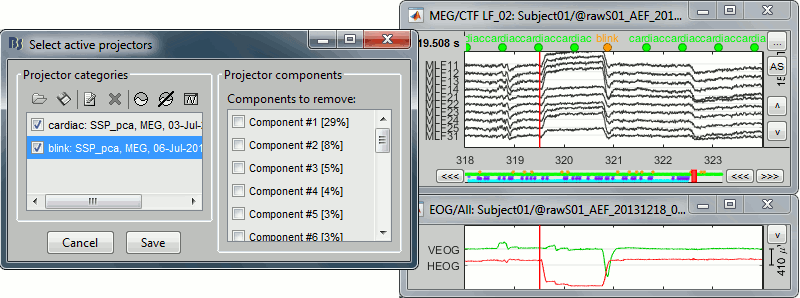
With only the component #2 selected (saccade):
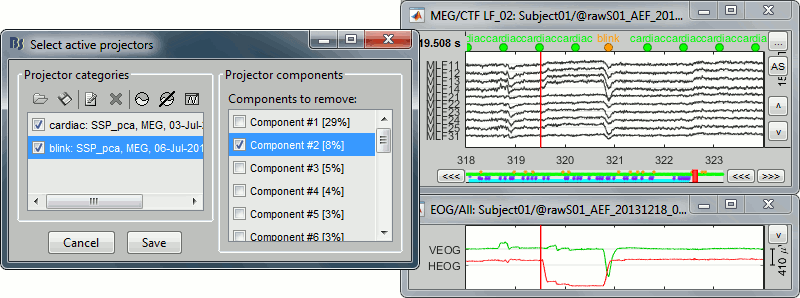
With components #1 and #2 selected (blink + saccade):
Keep the components #1 and #2 selected and click on [Save] to validate your changes.
Run #02: Running from a script
Let's perform the same detection operations on Run #02, using this time the Process1 box.
- Close everything with the [X] button at the top-right corner of the Brainstorm window.
Select the run AEF #02 in the Process1 box, then select the following processes:
Events > Detect heartbeats: Select channel ECG, check "All file", event name "cardiac".
Events > Detect eye blinks: Select channel VEOG, check "All file", event name "blink".
Events > Remove simultaneous: Remove "cardiac", too close to "blink", delay 250ms.
Always check visually the results of the SSP cleaning executed from a script.
- Open the MEG sensors for Run #02.
In the Record tab, menu Artifacts > Select active projectors.
SSP: Generic
The calculation of the SSP for the heartbeats and the eye blinks are shortcuts to a more generic algorithm "Compute SSP: Generic". We do not need it here, but the illustration of its properties is interesting to understand the correction of ocular and cardiac artifacts:
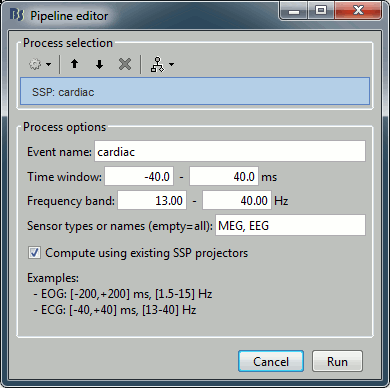
Event name: Markers that are used to characterize the artifact
Time window: Time segment to consider before and after each event marker. We want this time window to be longer than the artifact effect itself, in order to have a large number of time samples representative of a normal brain activity. This helps the SVD decomposition to separate the artifact from the ongoing brain activity.
Frequency band: Definition of the band-pass filter that is applied to the recordings before calculating the projector. Usually you would use the same frequency band as we used for the detection, but you may want to try to refine this parameter if the results are not satisfying.
Sensor types or names: List of sensors for which to calculate the projector. You can get better results if you process one sensor type at a time.
Evaluation
Eye blinks
One efficient way of representing the impact of this artifact correction is to epoch the recordings around the artifacts before and after the correction. Use the same time window as the one used in the SSP options around each marker, and average all the segments of recordings. Those operations are detailed in the next tutorial, we are just presenting the results. You don't need to reproduce them at this time, just remember that it is doable, in case you need it with your own data. This is what the database would look like after those operations:
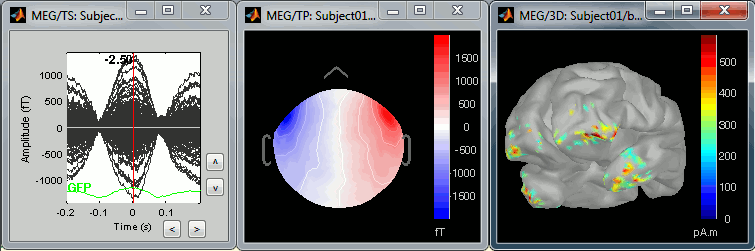
The following image represents the file "blink_uncorrected / Avg: blink", which is the average of the 18 identified blinks before the SSP correction, [-200,+200]ms around the "blink" events. The time series, the 2D topography at the peak (t = 0ms), and the mapping on the cortex of those fields, using the basic inverse model calculated in the previous tutorial.
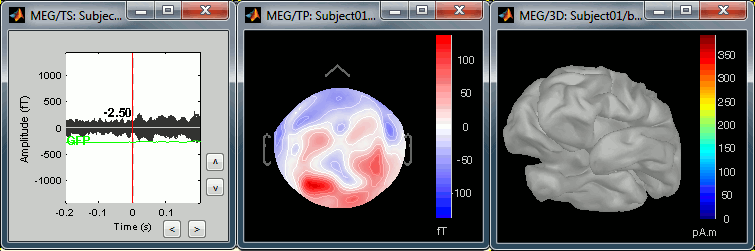
The next image represents the file "blink_ssp / Avg: blink", which is the exact same thing, but after the SSP correction. The artifact is gone. If you do this and you can still see some clear evoked component in the time series figure, the SSP correction was not efficient: the artifact is not properly identified, or you should select different components using the "Select active projectors" window.
Heartbeats
We can do the same thing with the heartbeats: epoch [-40,+40]ms around the "cardiac" events, average the trials, and review the event-related fields at three instants:
The peak of the P wave (t = -30ms):

The peak of the QRS complex (t = 0ms):
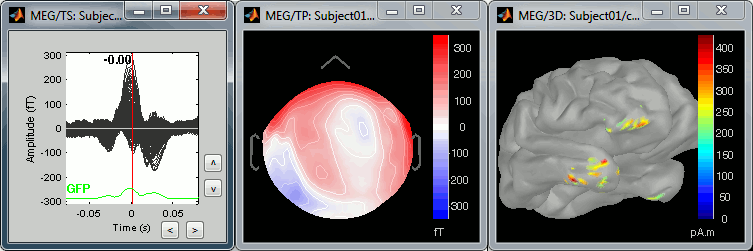
The peak of the T wave (t = 25ms):
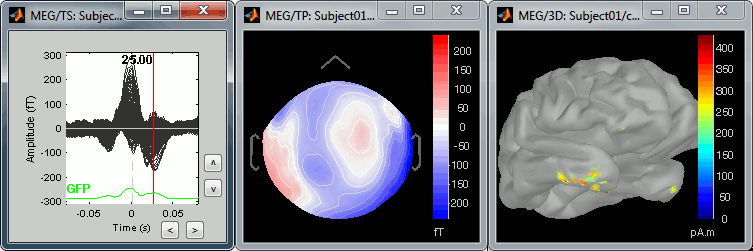
Those peaks may look big, but they are in fact much smaller (300 fT for the average of 346 repetitions) than what we observed for the eye blinks (1500 fT for 18 repetitions). You can notice that none of those topographies are similar to the components we obtained after calculating the SSP for the cardiac artifact. We don't know how to correct this artifact, it doesn't look too bad in terms of recordings contamination, so we just leave it uncorrected.
Troubleshooting
You have calculated the SSP as indicated here but you don't get any good results. No matter what you do, the topographies don't look like the targeted artifact. You can try the following:
- Review one by one the events indicating the artifacts, remove the ones that are less clear or that occur close to another artifact.
- Select or unselect the option "Compute using existing SSP".
- Use the process "Compute SSP: Generic" and modify some parameters:
- Reduce the time window around the peak of the artifact.
- Use a narrower frequency band: especially the EOG, if the projectors capture some of the alpha oscillations, you can limit the frequency band to [1.5 - 9] Hz.
- Change the method: Average / SSP.
- If you have multiple acquisition runs, you may try to use all the artifacts from all the runs rather than processing the files one by one. For that, use the Process2 tab instead of Process1. Put the "Link to raw file" of all the runs on both side, Files A (what is used to compute the SSP) and Files B (where the SSP are applied).
Always look at what this procedure gives you in output. Most of the time, the artifact cleaning will be an iterative process where to need several time to adjust the options and the order of the different steps in order to get good results.
Where are the projectors stored?
Theory
The Signal-Space Projection (SSP) is one approach to rejection of external disturbances. Here is a short description of the method by Matti Hämäläinen, from the MNE 2.7 reference manual, section 4.16.
Unlike many other noise-cancellation approaches, SSP does not require additional reference sensors to record the disturbance fields. Instead, SSP relies on the fact that the magnetic field distributions generated by the sources in the brain have spatial distributions sufficiently different from those generated by external noise sources. Furthermore, it is implicitly assumed that the linear space spanned by the significant external noise patterns has a low dimension.
Without loss of generality we can always decompose any n-channel measurement b(t) into its signal and noise components as:
b(t) = bs(t) + bn(t)
Further, if we know that bn(t) is well characterized by a few field patterns b1...bm, we can express the disturbance as
bn(t) = Ucn(t) + e(t) ,
where the columns of U constitute an orthonormal basis for b1...bm, cn(t) is an m-component column vector, and the error term e(t) is small and does not exhibit any consistent spatial distributions over time, i.e., Ce = E{eeT} = I. Subsequently, we will call the column space of U the noise subspace. The basic idea of SSP is that we can actually find a small basis set b1...bm such that the conditions described above are satisfied. We can now construct the orthogonal complement operator
P⊥ = I - UUT
and apply it to b(t) yielding
b(t) ≈ P⊥bs(t) ,
since P⊥bn(t) = P⊥Ucn(t) ≈ 0. The projection operator P⊥ is called the signal-space projection operator and generally provides considerable rejection of noise, suppressing external disturbances by a factor of 10 or more. The effectiveness of SSP depends on two factors:
The basis set b1...bm should be able to characterize the disturbance field patterns completely and
The angles between the noise subspace space spanned by b1...bm and the signal vectors bs(t) should be as close to π/2 as possible.
If the first requirement is not satisfied, some noise will leak through because P⊥bn(t) ≠ 0. If the any of the brain signal vectors bs(t) is close to the noise subspace not only the noise but also the signal will be attenuated by the application of P⊥ and, consequently, there might by little gain in signal-to-noise ratio.
Since the signal-space projection modifies the signal vectors originating in the brain, it is necessary to apply the projection to the forward solution in the course of inverse computations.
Algorithm
We have now 346 examples of heartbeats, 18 examples of blinks, and 12 examples of saccades. Those are sufficient numbers of repetitions to calculate reliable signal-space projections. The logic of the computation is the following:
- Take a small time window around each marker to capture the full effect of the artifact, plus some clean brain signals before and after. The default time window is [-200,+200]ms for eye blinks, and [-40,+40]ms for the heartbeats.
- Filter the MEG or EEG signals in a frequency band of interest, in which the artifact is the most visible.
Concatenate all those time blocks into a big matrix A = [b1, ..., bm]
Compute the singular value decomposition of this matrix A: [U,S,V] = svd(A, 'econ')
The singular vectors Ui with the highest singular values Si are an orthonormal basis of the artifact subspace that we want to subtract from the recordings. The software selects by default the vectors with eigenvalues above a certain threshold: Si > 12% * sum(Si)
Then it is possible to redefine interactively the selected components.Calculate the projection operator: P⊥i = I - UiUiT
Apply this projection on the MEG or EEG recordings F: F = P⊥iF
The process has to be repeated separately several times:
- for each sensor type (EEG, MEG magnetometers, MEG gradiometers) and
- for each artifact, starting with the one that leads to the strongest contamination in amplitude on the sensors (in the case: the eye blinks)
- If you have difficulties removing one artifact or the other, you may try to process them in a different order. You may also try removing some of the artifact markers in the case of co-occurring artifacts. If a lot of the blinks happen at the same time as heartbeats, you may end up calculating projectors that mix both effects, but that do not remove efficiently one or the other. In this case, remove all the markers that happen in the segments contaminated by multiple artifacts.
Steps #1 to #5 are done automatically by the processes "SSP > Compute SSP" in the Record tab: the results, the vectors Ui, are saved in the database link ("Link to raw file") and in the channel file.
Steps #6 and #7 are calculated on the fly when reading a block of recordings from the continuous file: when using the raw viewer, running a process a process on the continuous file, or importing epochs in the database.
Step #8 is the manual control of the process. Take some time to understand what you are trying to remove and how to do it. Never trust blindly any fully automated artifact cleaning algorithm, always check manually what is removed from the recordings, and do not give up if the first results are not satisfying.
References
For more information on the SSP method, please consult the following publications:
- C. D. Tesche, M. A. Uusitalo, R. J. Ilmoniemi, M. Huotilainen, M. Kajola, and O. Salonen, "Signal-space projections of MEG data characterize both distributed and well-localized neuronal sources," Electroencephalogr Clin Neurophysiol, vol. 95, pp. 189-200, 1995.
- M. A. Uusitalo and R. J. Ilmoniemi, "Signal-space projection method for separating MEG or EEG into components," Med Biol Eng Comput, vol. 35, pp. 135-40, 1997.
Additional documentation
McGill guidelines
- SSP cookbook
- Examples of topographies
- Alpha and cardiac overlapping?