Tutorial 14: Additional bad segments
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
We have already corrected the recordings for the artifacts at fixed frequencies (power lines) and for some standard and reproducible artifacts (heartbeats and blinks). There are many other possible sources of noise that can make the recordings unusable in our analysis. This tutorial introduces how to identify and mark these bad segments.
Contents
Manual inspection
It is very important to mark bad segments (noisy sections) of the recordings before running any fancy analysis. It may save you hours of repeated work later, when you discover after processing all your data that you have to redo everything because you have left some huge artifacts in the recordings.
In terms of quality control, there is no automatic method that will give you results as good as a manual screening of the recordings. We recommend you always take a few minutes to scroll through all your files to identify and tag all the noisy segments. Do this full screening after you're done with all the other pre-processing steps (filtering and SSP/ICA cleaning) to remove what has not been corrected with other techniques.
At the beginning, it is not easy to separate what is too noisy from what is acceptable. This is usually an iterative process: at the first attempt you guess, you average the trials and estimate the sources and finally realize that are some eye movements left that are masking your effect of interest. You have to delete everything, add some bad segments and try again. On the contrary, if you reject too much data at the beginning, you may not have enough trials to observe your effect correctly. The balance is not easy to find, but you'll get good at it quickly. Brainstorm offers tools to do these operations easily, so a few trials and errors are not too dramatic. Just make sure you check the quality of your data at every step of the analysis, so that you don't go too far in the wrong direction.
To review your recordings and check for major artifacts, you can for instance:
- Display all the sensors in columns, you don't need to see each sensor individually.
- Use a large time window (page duration = 5-20s).
- Increase the gain of the sensors to see clearly the abnormally large values.
Scroll using the shortcuts corresponding to buttons [<<<] and [>>>]: F3, Shift+F3, F4, etc.
- Unselect the autoscale option ([AS] button on the right side of the figure) so that the amplitude doesn't change every time you move to the next page, it will make the review easier.
When you identify something that doesn't look good:
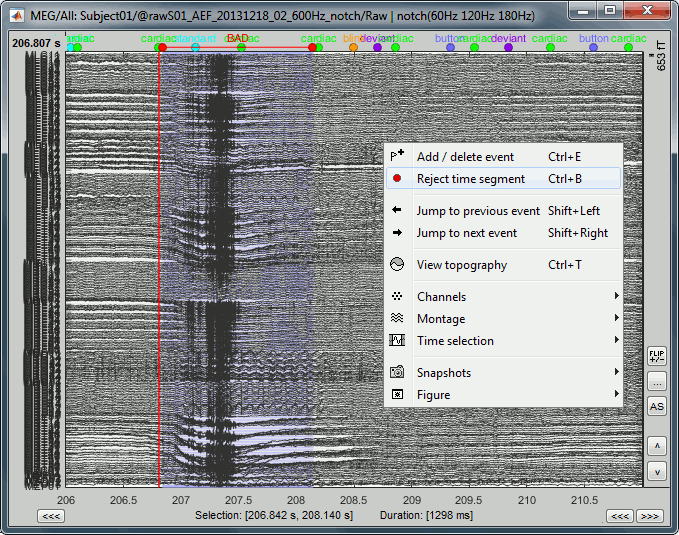
- Click and move your mouse to select the segment you consider as noisy.
Mark the segment as bad: Right-click > Reject time segment (or Ctrl+B).
The example below shows Run #02 from 206s to 211s:
Automatic detection
We have developed some tools to help with this screening procedure. The process "Artifacts > Detect other artifacts" identifies epochs of time that contain typical artifacts from eye movement, subject movement or muscle contractions. While it is still advised that you visually inspect all of your data, this process can help identify areas that contain artifacts which you may want to mark as bad segments.
Currently, the process runs only on continuous raw links and identifies artifacts in two frequency bands, chosen because of the predictability of artifacts in these bands.
1-7 Hz: Typically related to subject movement, eye movements and dental work (or other metal)
40-240 Hz: Related to muscle noise and some sensor artifacts
- Note that the alpha band (8-12 Hz) is specifically avoided here since some alpha oscillations can be quite high in amplitude and falsely detected as artifact.
Important notes
- Before running this detection it is highly recommended that you run the cleaning processes for cardiac and eye blink artifacts.
This process is currently being tested. If you find a bug or have other comments related to its performance, please provide comments here or on the user forum.
- We recommend you use the markers that this process creates as suggestions, not as the actual reality. Do not use this method fully automatically, always review its results.
Recommendations for usage
- Start by running the process on one run per subject. Scan though the recording and confirm that the detection is performing well.
- Adjust the threshold as needed, then run the detection on the other runs for that subject.
If there are many eye movements, the "1-7 Hz" detection can work well for computing an SSP projector. This is done using the menu "Artifacts > SSP: Generic" as described below. If a suitable projector is found and applied, re-run the artifact detection to find the remaining artifacts that were not removed.
Run #01
We will now apply this process on the first acquisition session:
Double-click on the link to show the MEG sensors for Run #01.
The process will exclude the segments that are already marked as bad from the detection. If you have marked bad segments at the beginning of this tutorial, delete them all: Select the category "BAD" and press the delete key, or menu Events > Delete group.
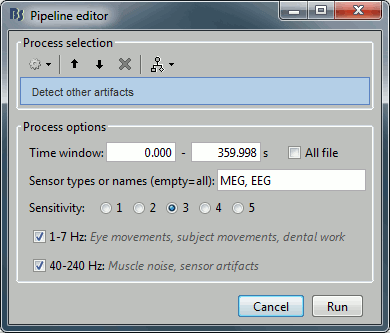
In the Record tab, select the menu: "Artifacts > Detect other artifacts".
Time window: The time window for which the detection will be performed.
Sensitivity: 1 is very sensitive, 5 very conservative, 3 works well for a variety of conditions.
Frequency band selection: Check the box for which band(s) you will perform the detection.
After running the process, event types are created, one for each frequency band. They contain extended events indicating the start and end of the epoch. The time resolution is 1 second and therefore the epoch may, in fact, be a bit longer than the actual artifact. You can manually refine the time definition if you wish and mark some or all events as bad.
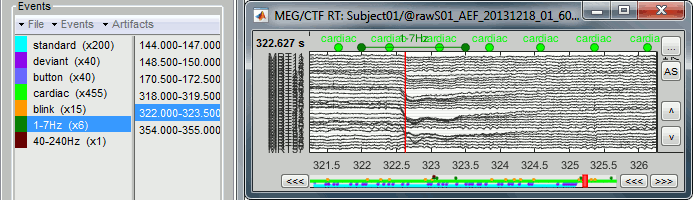
- Check quickly the segments that were marked as possible artifacts. They all correspond to some form of irregularities in the recordings but there are very few of them. We will just flag all the segments detected in both categories as bad.
For considering these events as bad, select them both in the events list and use the menu
"Events > Mark group as bad". Alternatively, you can rename the events and add the tag "bad_" in their name, it would have the same effect.
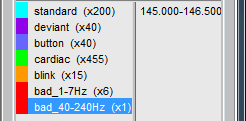
- Unload all the data with the [X] in the toolbar of the Brainstorm window. Save the modifications.
Run #02
Repeat the same operation on the second data file:
Double-click on the link to show the MEG sensors for Run #02.
Run the menu "Artifacts > Detect other artifacts" with the same parameters as the first file.
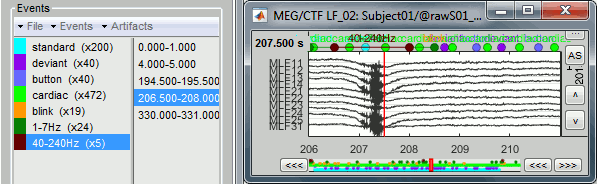
- The category "1-7Hz" contains many saccades, maybe enough for computing an SSP projector.
If you are not interested in seeing how to remove the saccades with SSP projectors, just mark the two groups as bad: with the menu "Events > Mark group as bad", or renaming them in "bad_1-7Hz" and "bad_40-240Hz". Then go directly to the next tutorial.
Saccade SSP
This run #02 is a good example to illustrate how we can use SSP projectors to remove the artifacts caused by eye saccades. You could mark the saccades manually or use the pre-selection available in "1-7Hz".
Rename category "1-7Hz" in "saccade" and delete category "40-240Hz".
Open the EOG recordings at the same time: right-click on file > EOG > Display time series.
Keep only the saccades: Delete all the events that do not show a clear step in the HEOG channel.
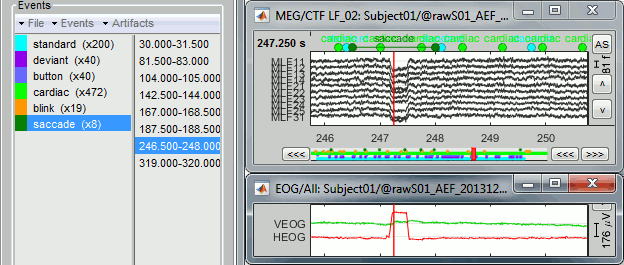
Run the process "Artifacts > SSP: Generic" with the following options:
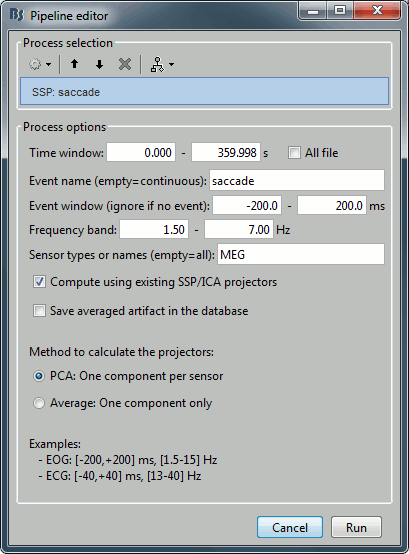
Note: the event window option will not be used because the events "saccade" are extended events and already include their duration.
The first component removes the artifact very well. Keep it selected and click on [Save].
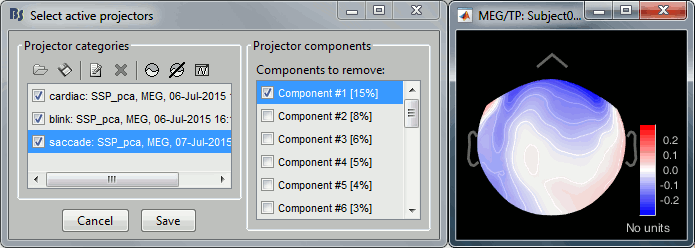
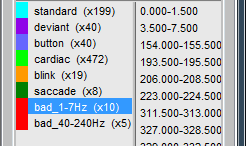
Run again the process "Detect other artifacts". There are now less events detected in 1-7Hz.
Rename the categories in "bad_1-7Hz" and "bad_40-240Hz" to flag them as bad segments.
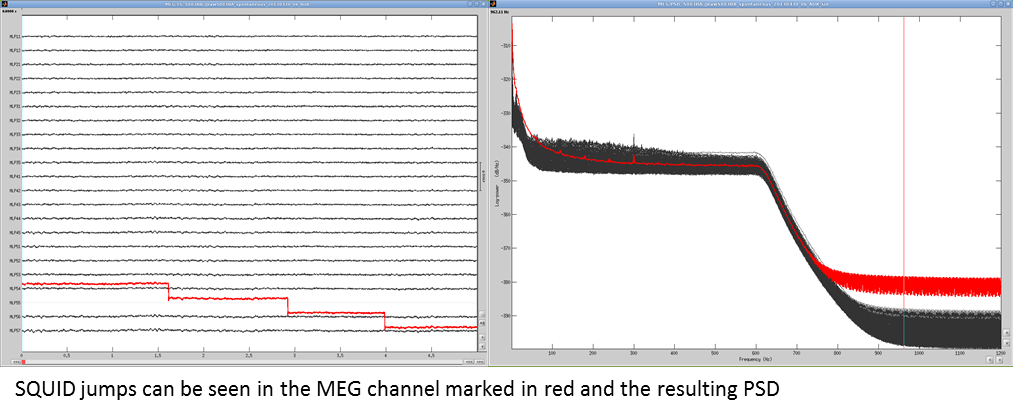
Elekta-Neuromag SQUID jumps
MEG signals recorded with Elekta-Neuromag systems frequently contain SQUID jumps. They are easy to spot visually in the recordings, they look like sharp steps followed by a change of baseline value. These jumps are due to the instability of the electronics, which fails at controlling the state of the SQUID during the recording sessions.
These steps cause important issues in the analysis of the signal, both in amplitude and in frequency. They are difficult to detect and remove, especially when some pre-processing with the Elekta software has already been applied. Running MaxFilter/SSS on MEG recordings with a SQUID jump on one sensor propagates the artifact to all the sensors.
The best approach is to remove these jumps from the analysis:
By marking them as bad segments manually, if their number is reasonable (in Brainstorm).
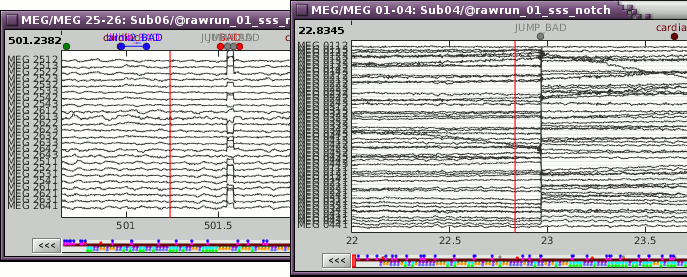
The 1-7Hz artifact detection employed here will usually catch them.By marking the sensors as bad if only a few of them are affected (before running MaxFilter).
By computing SSP projectors to remove the jumps (before running MaxFilter/SSS).
As of today, this approach is possible only if you use MNE or MNE-Python for the early stages of pre-processing. When the free SSS algorithm implemented in MNE-Python is made available in Brainstorm, we will be able to use this approach in Brainstorm as well.
An example before MaxFilter (SQUID jump visible on one sensor only):
Examples after MaxFilter (SQUID jump propagated on all the sensors):
Additional documentation
Tutorial: SSP cookbook