SEEG epileptogencity maps
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Olivier David.
This tutorial introduces some concepts that are specific to the management of SEEG recordings in the Brainstorm environment, and explains how to compute maps of epileptogenicity from ictal recordings. It is based on a clinical case from the Grenoble University Hospital, France.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Contents
Dataset description
License
This tutorial dataset (EEG and MRI data) remains property of the Grenoble University Hospital, France. Its use and transfer outside the ImaGIN and Brainstorm tutorials, e.g. for research purposes, is prohibited without written consent. For questions, please contact Olivier David, PhD ( olivier.david@inserm.fr ).
Clinical description
This dataset includes recordings for a patient that was not reported in the above article, but are part of the same study. The patient presents a focal epilepsy of the left temporo-occipital junction, MRI-negative, and was implanted in the surrounding areas. The subfolder "seeg" contains the recordings of three seizures, all of them showing a propagation of high-frequency oscillations from the lesion towards the temporal lobe, bilaterally.
SEEG recordings
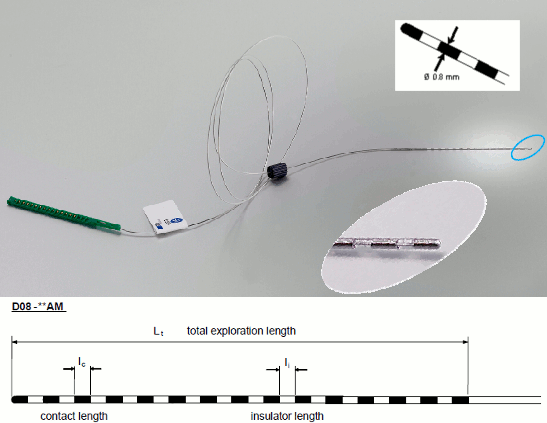
The depth electrodes used in this example dataset are DIXI D08-**AM Microdeep electrodes, with the following specifications:
- Diameter: 0.8 mm
- Contact length: 2 mm
- Insulator length: 1.5 mm
- Distance between the center of two contacts: 3.5 mm
- Between 8 and 18 contacts on each electrode
Files
The dataset we distribute with this tutorial follows the Brain Imaging Data Structure (BIDS) standard for neuroimaging data organization. This specification was first established for MRI and fMRI (Gorgolewski, 2016) and then refined with an extension dedicated to iEEG (Holdgraf, 2019). The files that will be imported in this tutorial are the following:
tutorial_epimap_bids/
derivatives/: Everything that cannot be considered as raw data
brainvisa/sub-01_ses-pre/: Result of the BrainVISA 4.5 segmentation for the pre-implantation T1 MRI.
sub-01/: Raw data for subject 01
ses-preimp/: Imaging exams performed before the implantation of the sEEG.
anat/sub-01_ses-preimp_T1w.nii.gz: T1-weighted MRI pre-implantation
ses-postimp/: Exams performed with the sEEG devices implanted.
anat/sub-01_ses-postimp_T1w.nii.gz: T1-weighted MRI post-implantation
ieeg/..._task-seizure_run-0*_ieeg.eeg: Three seizure recordings in BrainVision file format, one seizure per file (with the header files .vhdr and .vmrk)
ieeg/..._space-IXI549Space_electrodes.tsv: Position of the contacts in MNI space (SPM12 Segment non-linear normalization)
ieeg/..._space-ScanRAS_electrodes.tsv: Position of the contacts in world coordinates, relative to the post-implantation T1 MRI.
All the anatomical images have been de-identified with mri_deface from FreeSurfer 6.
References
The acquisition methodology is described in the following articles:
David O, Blauwblomme T, Job AS, Chabardès S, Hoffmann D, Minotti L, Kahane P, Imaging the seizure onset zone with stereo-electroencephalography, Brain. 2011 Oct;134(10):2898-911
Lamarche F, Job AS, Deman P, Bhattacharjee M, Hoffmann D, Gallazzini-Crépin C, Bouvard S, Minotti L, Kahane P, David O, Correlation of FDG-PET hypometabolism and SEEG epileptogenicity mapping in patients with drug-resistant focal epilepsy, Epilepsia. 2016 Dec; 57(12):2045–2055
Download and installation
How to get the example dataset:
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: tutorial_epimap.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
Files included in this package:
- anat/MRI/3DT1pre_deface.nii: Subject MRI before SEEG implantation
- anat/MRI/3DT1post_deface.nii: Subject MRI after SEEG implantation
- anat/MRI/brainvisa: Cortical surface extracted with BrainVISA 4.5
- anat/implantation/*: Positions of the SEEG contacts in various formats (MNI or subject space)
- seeg/SZ*.TRC: Seizure recordings in Micromed format, one seizure per file
MRI scans were de-identified with FreeSurfer's mri_deface.
Import the anatomy
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialEpimap" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
- Switch to the "anatomy" view of the protocol.
Right-click on the TutorialEpimap folder > New subject > Subject01
- Keep the default options you defined for the protocol.
Right-click on the subject node > Import MRI:
- Set the file format: "All MRI file (subject space)"
Select the file: tutorial_epimap/anat/MRI/3DT1pre_deface.nii
Do you want to apply the transformation to the MRI file? YES
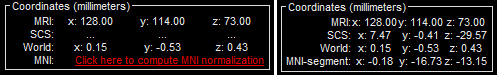
The MRI viewer opens automatically. Click on "Click here to compute MNI transformation". It computes an affine transformation between the subject space and the MNI ICBM152 space, and sets default positions for all the anatomical landmarks.