|
Size: 1273
Comment:
|
Size: 9660
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| Tuts #1-5 should have been done by the user | = Tutorial: Import and visualize functional NIRS data = ||<40%> || <!> ~+'''This tutorial is under construction'''+~ <!> || |
| Line 3: | Line 4: |
| Presentation of the experiment: * finger tapping + description | |
| Line 5: | Line 5: |
| * Aude's data: 2 runs of XXX minutes each * MRI anatomy 3T * link to dataset |
''Authors: Thomas Vincent, Zhengchen Cai'' |
| Line 9: | Line 7: |
| '''Import anatomy + MRI''' | The current tutorial assumes that the tutorials 1 to 6 have been performed. Even if they focus on MEG data, they introduce Brainstorm features that are required for this tutorial. |
| Line 11: | Line 9: |
| import Brainvisa head mesh and MRI file + provide datasets | List of prerequisites: |
| Line 13: | Line 11: |
| mark fudicials | * [[http://neuroimage.usc.edu/brainstorm/Tutorials/CreateProtocol|Create a new protocol]] * [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|Import the subject anatomy]] * [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy|Display the anatomy]] * [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile|Channel file / MEG-MRI coregistration]] * [[http://neuroimage.usc.edu/brainstorm/Tutorials/ReviewRaw|Review continuous recordings]] |
| Line 15: | Line 17: |
| '''Import NIRS functional data''' | == Download == The dataset used in this tutorial is available online. |
| Line 17: | Line 20: |
| NIRS Acquisition + description | * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and download the file: nirs_sample.zip * Unzip it in a folder that is not in any of the Brainstorm folders |
| Line 19: | Line 23: |
| dataset files: | == Presentation of the experiment == * Finger tapping task: 10 stimulation blocks of 30 seconds each, with rest periods of ~30 seconds * One subject, one NIRS acquisition run of 12''' '''minutes at 10Hz * 4 sources and 12 detectors (+ 4 proximity channels) placed above the right motor region * Two wavelengths: 690nm and 830nm * MRI anatomy 3T from /!\ ''' scanner type ''' |
| Line 21: | Line 30: |
| * 2 nirs files with proper SrcPos and DetPos fields | == Create the data structure == Create a protocol called "TutorialNIRSTORM": |
| Line 23: | Line 33: |
| -> generate these from brainsight output merged with montage coords | * Got to File -> New Protocol * Use the following setting : * '''Default anatomy''': Use individual anatomy. * '''Default channel file''': Use one channel file per subject (EEG). |
| Line 25: | Line 38: |
| -> check if there is a way to directly have this from brainsight without having to forge it | In term of sensor configuration, NIRS is very similar to EEG and the placement of optodes may change from subject to the other. |
| Line 27: | Line 40: |
| * 1 file of digitized coords of fiducials (NAS, LE, RE) as a txt file. Coords should be in the same referential as SrcPos and DetPos | (!) Should we add (EEG '''or NIRS''') in the interface? |
| Line 29: | Line 42: |
| '''NIRS-MRI coregistration''' | Create a subject called "Subject01" (Go to File -> New subject), with the default options |
| Line 31: | Line 44: |
| Use automatic registration | == Import anatomy == === Import MRI and meshes === Make sure you are in the anatomy view of the protocol. |
| Line 33: | Line 48: |
| Keep only step 1 | Right-click on "Subject01 -> Import anatomy folder". Select anatomy folder from the nirs_sample data folder. Reply "yes" when asked to apply the transformation. Leave the number of vertices for the head mesh to the default value. |
| Line 35: | Line 50: |
| Display Optodes (adapted from “Display Sensors”) | This will open the MRI review panel where you have to set the fudicial points (See [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|Import the subject anatomy]]). |
| Line 37: | Line 52: |
| - no helmet here -> should be able to show optode positions over head meshSKip manual registration | Note that the PC, AC and IH points are already defined. |
| Line 39: | Line 54: |
| Edit the channel file | {{attachment:NIRSTORM_tut1_MRI_edit_v2.gif||height="335"}} |
| Line 41: | Line 56: |
| Introduce new nomenclature | Here are the MRI coordinates (mm) of the fudicials used to produce the above figure: |
| Line 43: | Line 58: |
| S1D1WL1 NIRS_WL1 …S1D1WL2 NIRS_WL2 … | * NAS: x:95 y:213 z:114 * LPA: x:31 y:126 z:88 * RPA: x:164 y:128 z:89 * AC: x:96 y:137 z:132 * PC: x:97 y:112 z:132 * IH: x:95 y:103 z:180 |
| Line 45: | Line 65: |
| '''Visualize signals ''' | The head and white segmentations provided in the NIRS sample data were computed with Brainvisa and should automatically be imported and processed. |
| Line 47: | Line 67: |
| Depends on tut #5 | You can check the registration between the MRI and the loaded meshes by right-clicking on each mesh element and going to "MRI registration -> Check MRI/Surface registration". |
| Line 49: | Line 69: |
| Introduce default channel groups: ALL, WL1, WL2 | {{attachment:NIRSTORM_tut1_new_MRI_meshes.gif||height="335",width="355"}} |
| Line 51: | Line 71: |
| Just adapt sections “Montage selection” and “Channel selection” | == Import NIRS functional data == The functional data used in this tutorial was produced by the Brainsight acquisition software and is available in the data subfolder of the nirs sample folder. It contains the following files: * '''fiducials.txt''': the coordinates of the fudicials (nasion, left ear, right ear).<<BR>>The positions of the Nasion, LPA and RPA should have been digitized at the same location as the fiducials previously marked on the anatomical MRI. These points will be used by Brainstorm for the registration, hence the consistency between the digitized and marked fiducials is essential for good results. * '''optodes.txt''': the coordinates of the optodes (sources and detectors), in the same referential as in fiducials.txt. Note: the actual referential is not relevant here, as the registration will be performed by Brainstorm afterwards. * '''S01_Block_FO_LH_Run01.nirs''': the NIRS data in a HOMer-based format /!\ '''document format'''.<<BR>>Note: The fields ''SrcPos'' and ''DetPos'' will be overwritten to match the given coordinates in "optodes.txt" To import this data set in Brainstorm: * Go to the "functional data" view of the protocol. * Right-click on "Subject01 -> Review raw file" * Select file type "NIRS: Brainsight (.brs)" * Load the file "S01_Block_FO_LH_Run01.nirs" in the NIRS sample folder.<<BR>>Note: the importation process assumes that the files optodes.txt and fiducials.txt are in the same folder<<BR>>as the .nirs data file. == Registration == In the same way as in the tutorial "[[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile|Channel file / MEG-MRI coregistration]]", the registration between the MRI and the NIRS is first based on three reference points Nasion, Left and Right ears. It can then be refined with the either the full head shape of the subject or with manual adjustment. === Step 1: Fiducials === * The initial registration is based on the three fiducial point that define the Subject Coordinate System (SCS): nasion, left ear, right ear. You have marked these three points in the MRI viewer in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/NIRSDataImport#Import_MRI|previous part]]. * These same three points have also been marked before the acquisition of the NIRS recordings. The person who recorded this subject digitized their positions with a tracking device (here Brainsight). The position of these points are saved in the NIRS datasets (see fiducials.txt). * When we bring the NIRS recordings into the Brainstorm database, we align them on the MRI using these fiducial points: we match the NAS/LPA/RPA points digitized with Brainsight with the ones we placed in the MRI Viewer. * This registration method gives approximate results. It can be good enough in some cases, but not always because of the imprecision of the measures. The tracking system is not always very precise, the points are not always easy to identify on the MRI slides, and the very definition of these points does not offer a millimeter precision. All this combined, it is easy to end with an registration error of 1cm or more. * The quality of the source analysis we will perform later is highly dependent on the quality of the registration between the sensors and the anatomy. If we start with a 1cm error, this error will be propagated everywhere in the analysis. === Step 2: Head shape === /!\ We don't have digitized head points for this data set. We should skip this === Step 3: manual adjustment === /!\ TODO? To review this registration, right-click on "Common files / NIRS sensors (96) -> Display sensors" Show the fiducials, which were stored as additional digitized head points: right-clicj on "Common files / NIRS Sensors (96) -> Digitized head points -> View head points" {{attachment:NIRSTORM_tut1_display_sensors_fiducials.png||height="355"}} /!\ TOFIX: the registration is buggy /!\ TOFIX: only the sources are displayed -> also show detectors As reference, the following figures show the position of fiducials [blue] (inion and nose tip are extra positions), sources [orange] and detectors [green] as they were digitized by Brainsight: {{attachment:NIRSTORM_tut1_brainsight_head_mesh_fiducials_1.gif||height="280"}} {{attachment:NIRSTORM_tut1_brainsight_head_mesh_fiducials_2.gif||height="280"}} {{attachment:NIRSTORM_tut1_brainsight_head_mesh_fiducials_3.gif||height="280"}} == Review Channel information == The resulting data organization should be: {{attachment:NIRSTORM_tut1_func_organization.png}} This indicates that the data comes from the Brainsight system (BRS) and comprises 96 channels. To review the content of channels, right-click on "BS channels -> Edit channel file". {{attachment:NIRSTORM_tut1_channel_table.png||height="250"}} * /!\ TODO add: the channel named "STIM1" encodes the stimulation paradigm * Channels contain the NIRS time-series measurements. For a given NIRS channel, its name is composed of the pair Source / Detector and the wavelength value. Column Loc(1) contains the coordinates of the source, Loc(2) the coordinates of the associated detector. * Each NIRS channel is assigned to the group "NIRS_WL690" or "NIRS_WL830" so specified its wavelength. * The comment "NIRS_PROX" indicates that the channel is a close-source measurement.<<BR>>TODO: keep NIRS_PROX ?? == Visualize NIRS signals == Select "Subject01/S01_Block_FO_LH_Run01/Link to raw file -> NIRS -> Display time series" It will open a new figure with superimposed channels {{attachment:NIRSTORM_tut1_time_series_stacked.png||height="400"}} Which can also be viewed in butterfly mode {{attachment:NIRSTORM_tut1_time_series_butterfly.png||height="400"}} We refer to the tutorial for navigating in these views [[Tutorials/ReviewRaw|"Review continuous recordings"]] === Montage selection === /!\ TODO Within the NIRS channel type, the following channel groups are available: * '''All channels: '''gathers all NIRS channels * '''NIRS_WL690''': contains only the channels corresponding to the 690nm wavelength * '''NIRS_WL830''': contains only the channels corresponding to the 830nm wavelength * '''NIRS_PROXY''': contains only the proximity channels /!\ Add screenshot |
Tutorial: Import and visualize functional NIRS data
|
|
Authors: Thomas Vincent, Zhengchen Cai
The current tutorial assumes that the tutorials 1 to 6 have been performed. Even if they focus on MEG data, they introduce Brainstorm features that are required for this tutorial.
List of prerequisites:
Download
The dataset used in this tutorial is available online.
Go to the Download page of this website, and download the file: nirs_sample.zip
- Unzip it in a folder that is not in any of the Brainstorm folders
Presentation of the experiment
- Finger tapping task: 10 stimulation blocks of 30 seconds each, with rest periods of ~30 seconds
One subject, one NIRS acquisition run of 12 minutes at 10Hz
- 4 sources and 12 detectors (+ 4 proximity channels) placed above the right motor region
- Two wavelengths: 690nm and 830nm
MRI anatomy 3T from
 scanner type
scanner type
Create the data structure
Create a protocol called "TutorialNIRSTORM":
Got to File -> New Protocol
- Use the following setting :
Default anatomy: Use individual anatomy.
Default channel file: Use one channel file per subject (EEG).
In term of sensor configuration, NIRS is very similar to EEG and the placement of optodes may change from subject to the other.
![]() Should we add (EEG or NIRS) in the interface?
Should we add (EEG or NIRS) in the interface?
Create a subject called "Subject01" (Go to File -> New subject), with the default options
Import anatomy
Import MRI and meshes
Make sure you are in the anatomy view of the protocol.
Right-click on "Subject01 -> Import anatomy folder". Select anatomy folder from the nirs_sample data folder. Reply "yes" when asked to apply the transformation. Leave the number of vertices for the head mesh to the default value.
This will open the MRI review panel where you have to set the fudicial points (See Import the subject anatomy).
Note that the PC, AC and IH points are already defined.
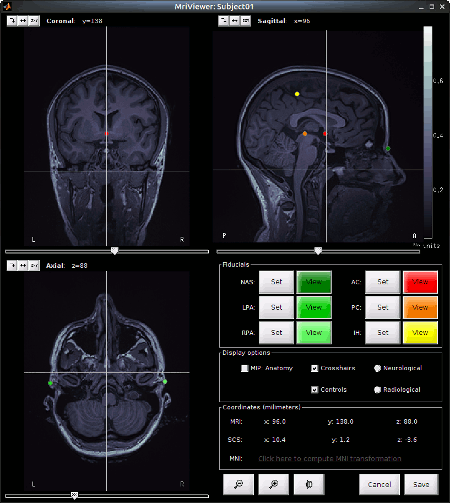
Here are the MRI coordinates (mm) of the fudicials used to produce the above figure:
- NAS: x:95 y:213 z:114
- LPA: x:31 y:126 z:88
- RPA: x:164 y:128 z:89
- AC: x:96 y:137 z:132
- PC: x:97 y:112 z:132
- IH: x:95 y:103 z:180
The head and white segmentations provided in the NIRS sample data were computed with Brainvisa and should automatically be imported and processed.
You can check the registration between the MRI and the loaded meshes by right-clicking on each mesh element and going to "MRI registration -> Check MRI/Surface registration".
Import NIRS functional data
The functional data used in this tutorial was produced by the Brainsight acquisition software and is available in the data subfolder of the nirs sample folder. It contains the following files:
fiducials.txt: the coordinates of the fudicials (nasion, left ear, right ear).
The positions of the Nasion, LPA and RPA should have been digitized at the same location as the fiducials previously marked on the anatomical MRI. These points will be used by Brainstorm for the registration, hence the consistency between the digitized and marked fiducials is essential for good results.optodes.txt: the coordinates of the optodes (sources and detectors), in the same referential as in fiducials.txt. Note: the actual referential is not relevant here, as the registration will be performed by Brainstorm afterwards.
S01_Block_FO_LH_Run01.nirs: the NIRS data in a HOMer-based format
 document format.
document format.
Note: The fields SrcPos and DetPos will be overwritten to match the given coordinates in "optodes.txt"
To import this data set in Brainstorm:
- Go to the "functional data" view of the protocol.
Right-click on "Subject01 -> Review raw file"
- Select file type "NIRS: Brainsight (.brs)"
Load the file "S01_Block_FO_LH_Run01.nirs" in the NIRS sample folder.
Note: the importation process assumes that the files optodes.txt and fiducials.txt are in the same folder
as the .nirs data file.
Registration
In the same way as in the tutorial "Channel file / MEG-MRI coregistration", the registration between the MRI and the NIRS is first based on three reference points Nasion, Left and Right ears. It can then be refined with the either the full head shape of the subject or with manual adjustment.
Step 1: Fiducials
The initial registration is based on the three fiducial point that define the Subject Coordinate System (SCS): nasion, left ear, right ear. You have marked these three points in the MRI viewer in the previous part.
- These same three points have also been marked before the acquisition of the NIRS recordings. The person who recorded this subject digitized their positions with a tracking device (here Brainsight). The position of these points are saved in the NIRS datasets (see fiducials.txt).
- When we bring the NIRS recordings into the Brainstorm database, we align them on the MRI using these fiducial points: we match the NAS/LPA/RPA points digitized with Brainsight with the ones we placed in the MRI Viewer.
- This registration method gives approximate results. It can be good enough in some cases, but not always because of the imprecision of the measures. The tracking system is not always very precise, the points are not always easy to identify on the MRI slides, and the very definition of these points does not offer a millimeter precision. All this combined, it is easy to end with an registration error of 1cm or more.
- The quality of the source analysis we will perform later is highly dependent on the quality of the registration between the sensors and the anatomy. If we start with a 1cm error, this error will be propagated everywhere in the analysis.
Step 2: Head shape
![]() We don't have digitized head points for this data set. We should skip this
We don't have digitized head points for this data set. We should skip this
Step 3: manual adjustment
![]() TODO?
TODO?
To review this registration, right-click on "Common files / NIRS sensors (96) -> Display sensors"
Show the fiducials, which were stored as additional digitized head points: right-clicj on "Common files / NIRS Sensors (96) -> Digitized head points -> View head points"
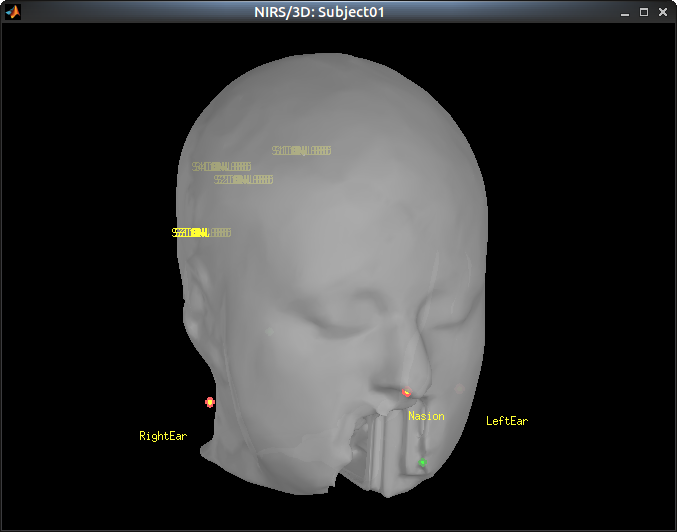
![]() TOFIX: the registration is buggy
TOFIX: the registration is buggy
![]() TOFIX: only the sources are displayed -> also show detectors
TOFIX: only the sources are displayed -> also show detectors
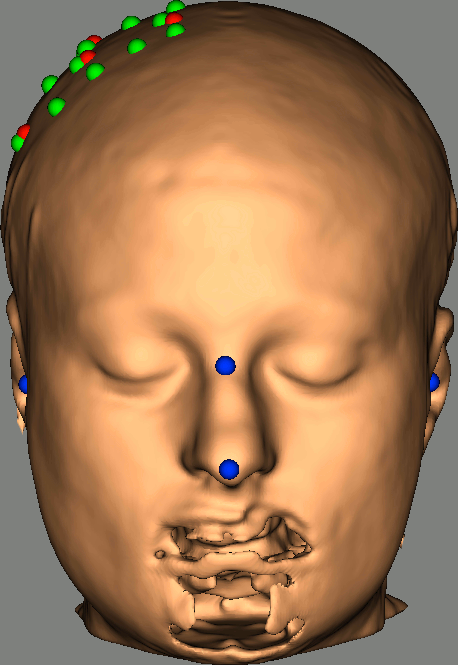
As reference, the following figures show the position of fiducials [blue] (inion and nose tip are extra positions), sources [orange] and detectors [green] as they were digitized by Brainsight:
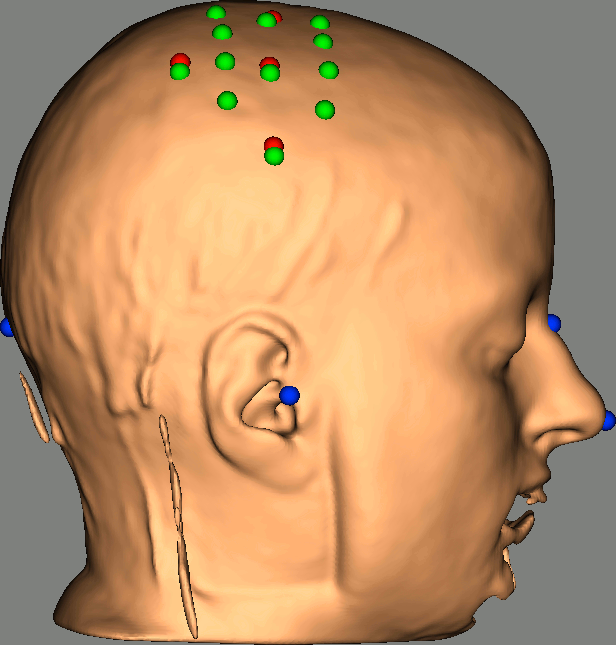
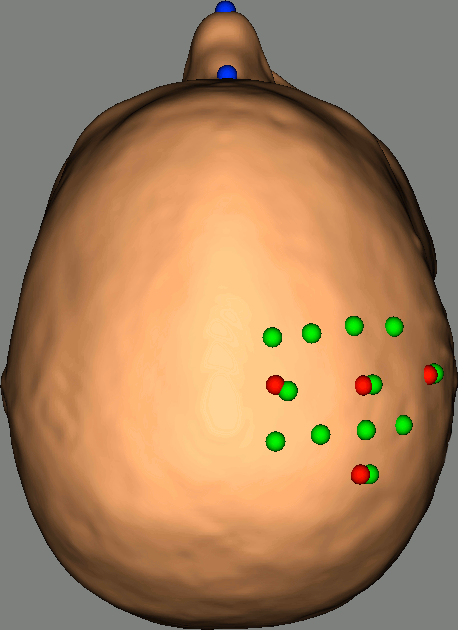
Review Channel information
The resulting data organization should be:
This indicates that the data comes from the Brainsight system (BRS) and comprises 96 channels.
To review the content of channels, right-click on "BS channels -> Edit channel file".
 TODO add: the channel named "STIM1" encodes the stimulation paradigm
TODO add: the channel named "STIM1" encodes the stimulation paradigm - Channels contain the NIRS time-series measurements. For a given NIRS channel, its name is composed of the pair Source / Detector and the wavelength value. Column Loc(1) contains the coordinates of the source, Loc(2) the coordinates of the associated detector.
- Each NIRS channel is assigned to the group "NIRS_WL690" or "NIRS_WL830" so specified its wavelength.
The comment "NIRS_PROX" indicates that the channel is a close-source measurement.
TODO: keep NIRS_PROX ??
Visualize NIRS signals
Select "Subject01/S01_Block_FO_LH_Run01/Link to raw file -> NIRS -> Display time series"
It will open a new figure with superimposed channels
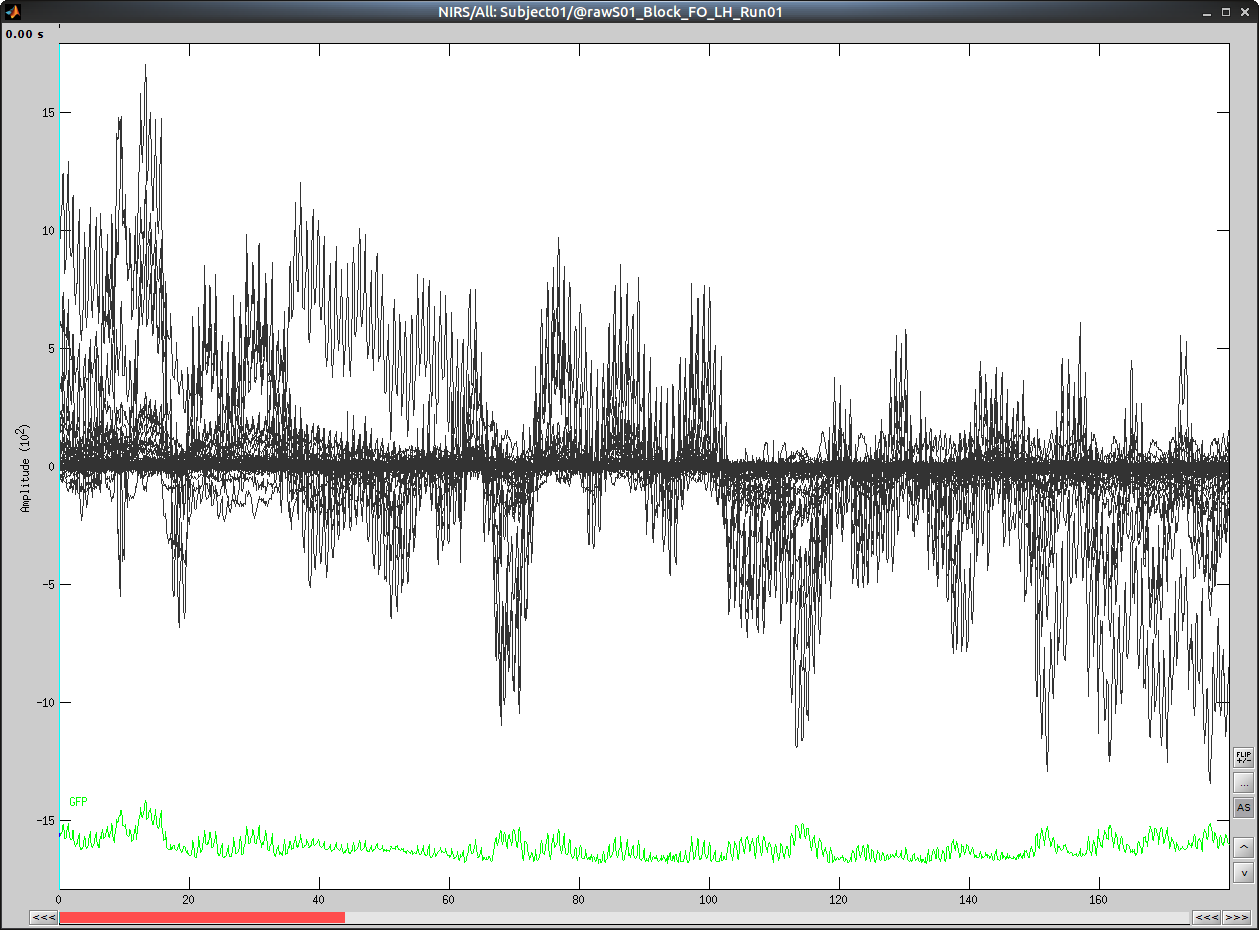
Which can also be viewed in butterfly mode
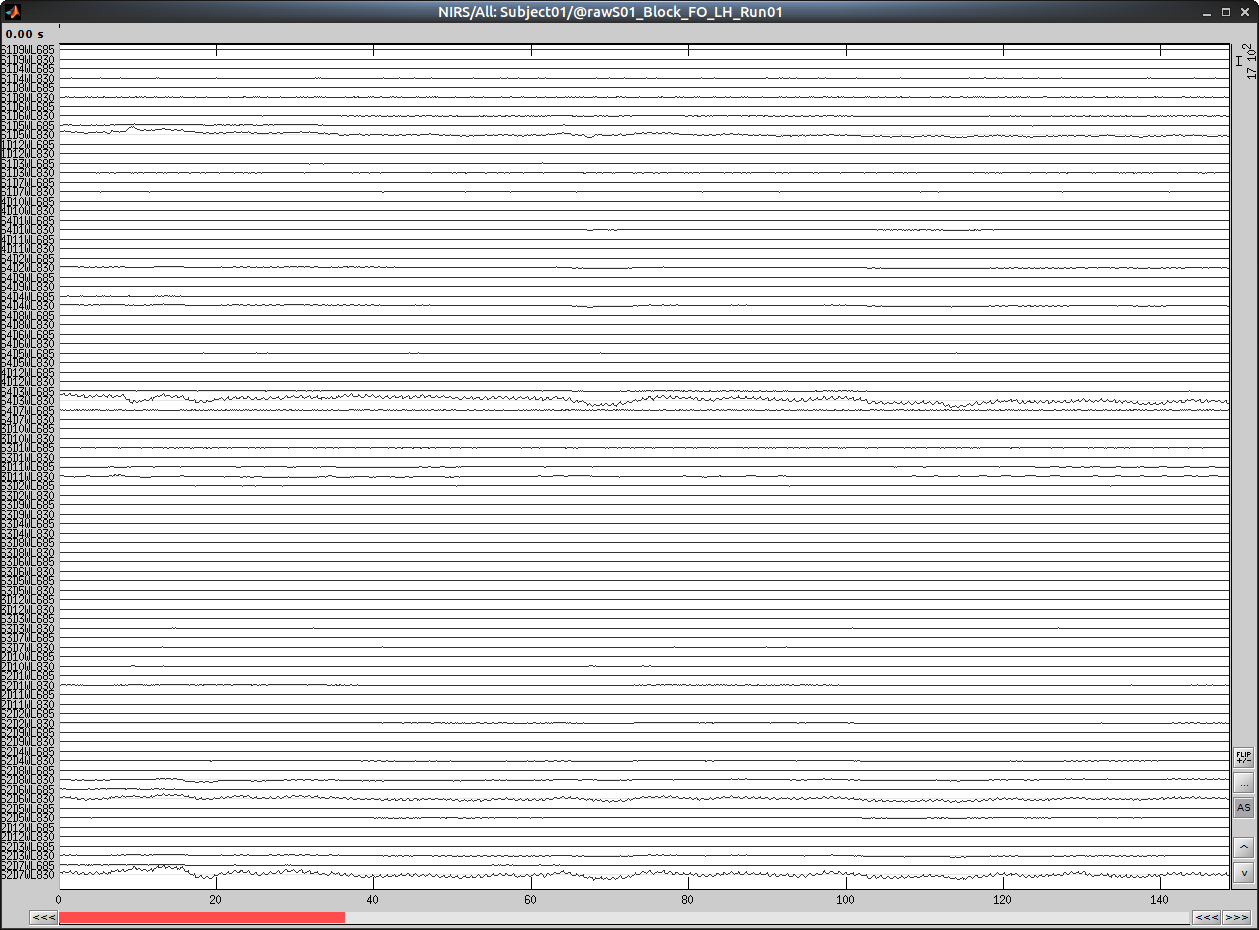
We refer to the tutorial for navigating in these views "Review continuous recordings"
Montage selection
![]() TODO
TODO
Within the NIRS channel type, the following channel groups are available:
All channels: gathers all NIRS channels
NIRS_WL690: contains only the channels corresponding to the 690nm wavelength
NIRS_WL830: contains only the channels corresponding to the 830nm wavelength
NIRS_PROXY: contains only the proximity channels
![]() Add screenshot
Add screenshot
