|
Size: 12765
Comment:
|
Size: 12610
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 5: Review continuous files = | = Tutorial 5: Review continuous recordings = |
| Line 6: | Line 6: |
| == Presentation of the experiment == A full description of this dataset is available on this page: [[http://neuroimage.usc.edu/brainstorm/DatasetAuditory|MEG auditory dataset]]. ==== Experiment ==== * One subject, two acquisition runs of 6 minutes each * Subject stimulated binaurally with intra-aural earphones (air tubes+transducers) * Each run contains: * 200 regular beeps (440Hz) * 40 easy deviant beeps (554.4Hz, 4 semitones higher) * Random inter-stimulus interval: between 0.7s and 1.7s seconds, uniformly distributed * The subject presses a button when detecting a deviant * Auditory stimuli generated with the Matlab Psychophysics toolbox ==== MEG acquisition ==== * Acquisition at '''2400Hz''', with a '''CTF 275''' system at the MNI, subject in seating position * Online 600Hz low-pass filter, files saved with the 3rd order gradient * Recorded channels (340): * 1 Stim channel indicating the presentation times of the audio stimuli: UDIO001 (#1) * 1 Audio signal sent to the subject: UADC001 (#316) * 1 Response channel recordings the finger taps in response to the deviants: UPPT001 (#2) * 26 MEG reference sensors (#5-#30) * 274 MEG axial gradiometers (#31-#304) * 2 EEG electrodes: Cz, Pz (#305 and #306) * 1 ECG bipolar (#307) * 2 EOG bipolar (vertical #308, horizontal #309) * 12 Head tracking channels: Nasion XYZ, Left XYZ, Right XYZ, Error N/L/R (#317-#328) * 20 Unused channels (#3, #4, #310-#315, #329-340) * 3 datasets: * '''S01_AEF_20131218_01.ds''': Run #1, 360s, 200 standard + 40 deviants * '''S01_AEF_20131218_02.ds''': Run #2, 360s, 200 standard + 40 deviants * '''S01_Noise_20131218_01.ds''': Empty room recordings, 30s long * File name: S01=Subject01, AEF=Auditory evoked field, 20131218=date(Dec 18 2013), 01=run * Use of the .ds, not the AUX (standard at the MNI) because they are easier to manipulate in FieldTrip ==== Head shape and fiducial points ==== * 3D digitization using a Polhemus Fastrak device driven by Brainstorm ('''S01_20131218_*.pos''') More information: [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutDigitize|Digitize EEG electrodes and head shape]] * The output file is copied to each .ds folder and contains the following entries: * The position of the center of CTF coils * The position of the anatomical references we use in Brainstorm: Nasion and connections tragus/helix, as illustrated [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems#Pre-auricular_points_.28LPA.2C_RPA.29|here]]. * Around 150 head points distributed on the hard parts of the head (no soft tissues) = From Continuous = == Open the file == Right-click on the data file > MEG (all) > Display time series. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=displayTsMenu.gif|displayTsMenu.gif|class="attachment"}} |
== Open the recordings == * Let's look first at the first file in the list: AEF#01 * Right-click on the Link to raw file. Below the first to menus, you have the list of channel types: * '''MEG''': 274 axial gradiometers * '''ECG''': 1 electrocadiogram, bipolar electrode across the chest * '''EOG''': 2 electrooculograms (vertical and horizontal) * '''Misc''': EEG electrodes Cz and Pz * '''ADC A''': Unused * '''ADC V''': Auditory signal sent to the subject * '''DAC''': Unused * '''FitErr''': Fitting error when trying to localize the three head localization coils (NAS, LPA, RPA) * '''HLU''': Head Localizing Unit, displacements in the three directions (x,y,z) for the three coils * '''MEG REF''': 26 reference sensors used for removing the environmental noise * '''Other''': Unused * '''Stim''': Stimulation channel, records the stim triggers generated by the Psychophysics toolbox * '''SysClock''': System clock, unused * Select > MEG > Display time series (or double-click on the file).<<BR>><<BR>> {{attachment:link_menu.gif||height="322",width="324"}} |
| Line 62: | Line 32: |
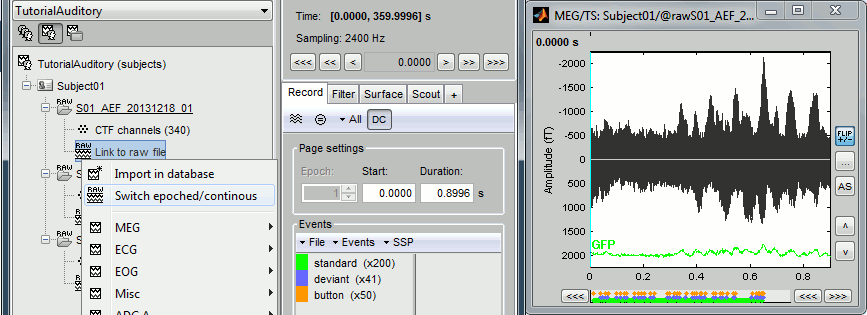
== Epoched vs. continuous == * The CTF MEG system can save two types of files: epoched (.ds) or continuous (_AUX.ds). * Here we have an intermediate storage type: continuous recordings saved in an "epoched" file. The file is saved as small blocks of recordings of a constant time length (1 second in this case). All those time blocks are contiguous, there is no gap between them. * Brainstorm can consider this file either as a continuous or an epoched file. By default it imports the regular .ds folders as epoched, but we can change this manually, to process it as a continuous file. * Double-click on the "Link to raw file" for run 01 to view the MEG recordings. You can navigate in the file by blocks of 1s, and switch between blocks using the "Epoch" box in the Record tab. The events listed are relative to the current epoch. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory?action=AttachFile&do=get&target=raw4.gif|raw4.gif|height="206",width="575",class="attachment"}} * Right-click on the "Link to raw file" for run 01 > '''Switch epoched/continuous''' * Double-click on the "Link to raw file" again. Now you can navigate in the file without interruptions. The box "Epoch" is disabled and all the events in the file are displayed at once. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory?action=AttachFile&do=get&target=raw5.gif|raw5.gif|height="209",width="576",class="attachment"}} * Repeat this operation twice to convert all the files to a continuous mode. * '''Run 02''' > Switch epoched/continuous * '''Noise''' > Switch epoched/continuous |
Tutorial 5: Review continuous recordings
Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet
Contents
Open the recordings
- Let's look first at the first file in the list: AEF#01
- Right-click on the Link to raw file. Below the first to menus, you have the list of channel types:
MEG: 274 axial gradiometers
ECG: 1 electrocadiogram, bipolar electrode across the chest
EOG: 2 electrooculograms (vertical and horizontal)
Misc: EEG electrodes Cz and Pz
ADC A: Unused
ADC V: Auditory signal sent to the subject
DAC: Unused
FitErr: Fitting error when trying to localize the three head localization coils (NAS, LPA, RPA)
HLU: Head Localizing Unit, displacements in the three directions (x,y,z) for the three coils
MEG REF: 26 reference sensors used for removing the environmental noise
Other: Unused
Stim: Stimulation channel, records the stim triggers generated by the Psychophysics toolbox
SysClock: System clock, unused
Select > MEG > Display time series (or double-click on the file).
You can see new information in the tab "Record" and a figure showing the recordings.
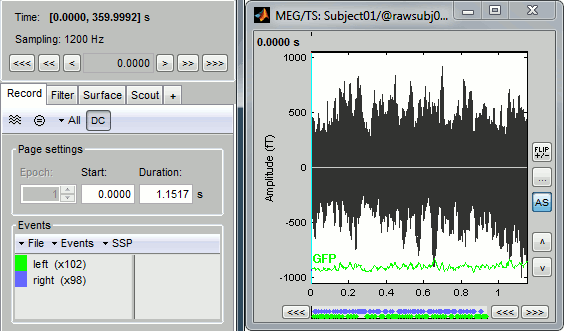
Navigate in time
As described in the basic tutorials, you can set the current time by using either the time panel (buttons and text field), or the figure (click on the white or grey areas of the figure). But you can notice that only a few seconds are visible in the figure, while the time panel (top left of the previous figure), indicates that we have 360s of recordings. Only a small block of the continuous file has been loaded in memory. This small time window can be configured with the tab Record/Page settings, with the text boxes Start and Duration.
The time series figure is similar to the ones that were presented in the previous tutorials, with a few new elements. The navigation bar at the bottom represents the time of the entire raw file, where the events are also represented by dots. The <<< and >>> buttons are the same as the ones in the time panel, and jump to the previous/next segment in the file. Clicking on the bar or dragging the red cursor change the current time window as well.
Epoched vs. continuous
- The CTF MEG system can save two types of files: epoched (.ds) or continuous (_AUX.ds).
- Here we have an intermediate storage type: continuous recordings saved in an "epoched" file. The file is saved as small blocks of recordings of a constant time length (1 second in this case). All those time blocks are contiguous, there is no gap between them.
- Brainstorm can consider this file either as a continuous or an epoched file. By default it imports the regular .ds folders as epoched, but we can change this manually, to process it as a continuous file.
- Double-click on the "Link to raw file" for run 01 to view the MEG recordings. You can navigate in the file by blocks of 1s, and switch between blocks using the "Epoch" box in the Record tab. The events listed are relative to the current epoch.
Right-click on the "Link to raw file" for run 01 > Switch epoched/continuous
- Double-click on the "Link to raw file" again. Now you can navigate in the file without interruptions. The box "Epoch" is disabled and all the events in the file are displayed at once.
- Repeat this operation twice to convert all the files to a continuous mode.
Run 02 > Switch epoched/continuous
Noise > Switch epoched/continuous
Channel selection
If you click on the lines instead of the white area: select channels.
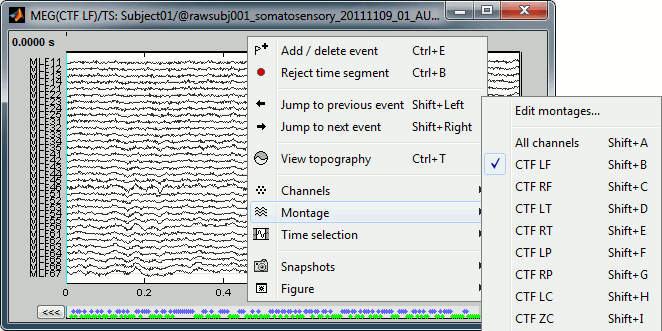
Montage selection
Let's switch to a nicer representation of the recordings time series: click on the "Display mode" button in the toolbar of the Record tab.
Now the traces are displayed in columns, but all the channels are displayed in the same figure, which makes it unreadable. Select a subset of channels by right-clicking on the figure > Montages, with the drop-down menu in the Record tab or with a keyboard shortcut (Shift+A, B, C...). Default groups of sensors are available for some MEG systems, but you can also create your own groups of sensors with the menu "Edit montages". The montage editor is described in the next tutorial.
Amplitude scale
In this display mode, the amplitude scale is represented on the right of the figure. You can adjust this vertical scale:
Use the buttons "^" and "v" on the right side of the figure. The shortcuts for those buttons are indicated in the tooltips (leave the mouse for a short while over a button)
Hold the Shift key and move the mouse wheel, or use the keys "+" and "-".
Use the button "..." on the right side of the figure ("Set scale manually") to set the scale to a precise level.
When scrolling in time to a different page, the amplitude scale is by default kept. You can change this behavior to re-evaluate automatically an optimal scale each time you change the current time window. This option is called "Auto-scale amplitude" and is disabled by default. To activate it: click on the "AS" button on the right of the figure, or check the menu "Display > Auto-scale amplitude" in the Record tab.
Display options
Remove DC offset: Button [DC] in the Record tab. When selected, for each channel, the average value over the entire current time window is subtracted from the channel values. This means that if you change the length of the time window, the value that is removed from each channel may change. It doesn't make much sense to disable this option for unprocessed MEG recordings.
Apply CTF compensation: Button [CTF] in the Record tab. Enable/disable the CTF noise correction based on the reference sensors, when it is not already applied in the file. In the current file, the CTF 3rd order gradient compensation is already applied, therefore this option is not available.
Flip +/-: Button in the right part of the time series figure. Exchange the direction of the Y axis, useful mostly for clinical EEG.
Set scale manually: Button [...] in the figure. Forces a defined amplitude scaling.
Auto-scale amplitude: Button [AS] in the figure. When selected, the vertical scale is adapted to the maximum value over the time window when the time window changes. When not selected: the vertical scales keeps its last value when you jump to another part of the file.
Online filter
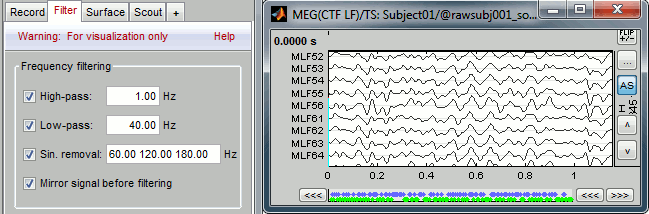
With the Filter tab, you can apply a band-pass filter to the recordings, or remove a set of specific frequencies (example: the 50Hz or 60Hz power lines contamination and their harmonics). The filters are applied only to the time window that is currently loaded; hence if the segment is too short for the required filters, the results could be inaccurate.
The option "Mirror signal before filtering" triples artificially the length of the signal with a mirror symmetry on each side, to avoid the strong edge effects that those filters can generate. Those online filters are not very accurate, they just provide a quick estimate for visualization only, the results are not saved anywhere. To filter properly the continuous files, please use the Process1 tab.
After testing the high-pass, low-pass and notch filters, uncheck them. If not you will probably forget about them, and they will stay on until you restart Brainstorm.
Shortcut summary
Keyboard shortcuts
Left / right arrows:
- No other key: Change current time, sample by sample
With Control key: Jump to previous/next time segment (same as the "<<<" and ">>>" buttons)
With Shift key: Jump to next event of the selected group
On MacOS, these shortcuts are different: please read the tooltips from the buttons ">", ">>", and ">>>" in the time panel to get the appropriate shortcuts.
Page-up / page-down:
- Same as left/right arrows, but faster (10 samples at a time)
If epochs are defined in the file: Control + page-up/page-down jumps to the next/previous epoch.
F3/Shift+F3: Jump to the next/previous epoch or page
F4/Shift+F4: Jump to the next/previous half-page
Plus / minus: Adjust the vertical scale of the time series
Control + E: Add / delete event occurrence
Control + T: Open a 2D topography window at the current time
Shift + Letter: Changes the set of electrodes currently displayed in the figure (list available by right-clicking on the figure > Display setup > ...)
Enter: Display the selected channels in a separate figure (selected channels = lines on which you clicked, that are shown in red)
Escape: Unselect all the selected channels
Delete: Mark the selected channels as bad
Mouse shortcuts
Mouse click on a channel: Select the channel
Mouse click: Change current time
Mouse click + Shift: For the selection of the current time (do not select any sensor, even when clicking on a line)
Mouse click + move: Select time range
Mouse wheel: Zoom around current time
Control + mouse wheel: Zoom vertically
Shift + mouse wheel: Adjust the vertical scale of the time series
Right-click: Display popup menu
Right-click + move: Move in a zoomed figure
Double click: Restore initial zoom settings (but do not restore the vertical scale of the time series)
From CTF
Time selection
Go back to the previous view mode ("butterfly view") and in the Record tab, select the menu "All channels" again. Keep this selection for the rest of the tutorial.
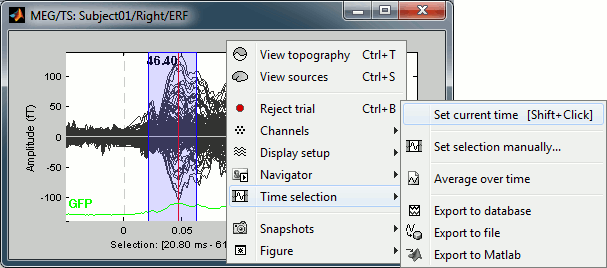
- Left-click somewhere on the white part of the time series figure, hold the mouse button, and drag your mouse left or right: A transparent blue rectangle appears to represent the time selection.
- Now right-click on the figure to see the functions you can apply on this time window. The selected time window and its duration are shown at the bottom of the figure.
Set current time: Just sets the time cursor when the right-click occurred. The shortcut Shift+Click can be useful when trying to move in time on dense displays in columns view.
Set selection manually: Select a time window by giving the bounds in ms.
Average over time: Average over the selected time window, saved as a new file in the database.
Export to database: Extract the recordings and save them in a new file in database.
- If some sensors are selected, only their values are extracted, all the others are set to zero.
Export to file: Same, but in a user-defined file (not in the database)
Export to Matlab: Same, but export as a Matlab variable in the current workspace.