Yokogawa/KIT tutorial
Authors: Francois Tadel, Yasuhiro Haruta, Ei-ichi Okumura, Takashi Asakawa
This tutorial introduces some concepts that are specific to the management of MEG/EEG files recorded with Yokogawa/KIT systems in the Brainstorm environment.
Contents
License
This tutorial dataset (MEG/EEG and MRI data) remains proprietary of Yokogawa Electric Corporation, Japan. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from Yokogawa Electric Corporation.
Description of the experiment
This tutorial is based on a simple median nerve stimulation experiment:
- Right median nerves were percutaneously stimulated using monophasic square-wave impulses with a duration of 0.3 ms at 2.8 Hz.
- The stimulus intensity was set at the motor threshold to evoke mild twitches of the thumb.
- The stimulus onsets were recorded as low-to-high TTL with a trigger channel labeled as "Trigger01".
- The total number of stimuli in the dataset was 336.
- The MEG data was recorded with a sampling rate of 2000 Hz and a bandpass filter at 0.16-500 Hz with a Yokogawa 160 axial gradiometer system at Yokogawa Electric Corporation, Kanazawa, Japan.
- The EEG data was recorded with a NIHON KOHDEN system simultaneously with the MEG recordings.
Export recordings from Meg160
To import Yokogawa/KIT data files (.con, .raw, .ave) into Brainstorm, a data export process is required beforehand. The data export function is available in Meg160, which is data analysis software equipped in most of Yokogawa/KIT systems.
The dataset used in this tutorial has already been exported using this procedure. It is described here so that later you can export your own recordings to Brainstorm.
If your software does not support the functions used below, please contact Yokogawa via
http://www.yokogawa.com/me/index.htm
Export the digitizer file
- If a wave data file and the corresponding digitizer file are ready for use, there is no additional operation required.
- If no digitizer file is available, you need first to extract the head surface points:
In Meg160, select the menu: File > Import and Export > BESA Text Export > Surface Point File
- Check that the fiducial points are properly pointed, and then click the [OK] button
- A surface point file (.sfp) is automatically created. It includes the position data of: the fiducial points, the marker points and other points describing the head shape
Full head shape in the the digitizer file
In order to realize a precise MRI registration or for warping the default anatomy (see tutorial), you should collect 100 to 200 points describing the entire head shape in addition to the 8 Yokogawa/KIT standard stylus points. To import additional digitized points, follow the instruction below:
- When digitizing head points:
- Pick the 8 standard stylus points
- Pick additional 100 to 200 head points, so that the selected points cover the entire head
Edit the digitizer label file (DigitizeLabel.txt) which is used in "Third-party export" so that it defines the 8 points and the additional points.
Note that the number of additional labels should be >= the number of digitized points.
- As for the new label names, any names can be used if the names are not duplicated.
- The following is an example:
fidt9 HPI_1 HPI_4 HPI_3 HPI_5 HPI_2 fidt10 fidnz ETC001 ETC002 ETC003 ... ETC[nnn]
Where [nnn]+8 should be >= the number of digitized points.
- Use the modified digitizer label file with the digitizer data in the "Third-party export" function.
Export the recordings
In Meg160, select the menu: File > Import and Export > Third-Party Export
- On some systems, this menu is name [BESA Binary Export]
- An operation panel for data export shows up
- When using digitizer data:
- Select [Digitizer]
Enter the digitizer file in the [Point Filename] box
(.txt file generally available under the corresponding "Scan" folder)Enter the label file in the [Label Filename] box
(DigitizeLabel.txt generally located in the "C:\Meg160\AppInfo" folder)
- When using surface point data instead:
- Select [Surface Point File]
- Enter the surface point file (.sfp) created previously.
- Enter an output file name in [Third-party Export Dataset]
- Click on [Create Export File]
- Use this exported file in Brainstorm
Download and installation
- Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: sample_yokogawa.zip
Unzip it in a folder that is not in any of the Brainstorm folders (program or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialYokogawa" and select the options:
"No, use individual anatomy",
"No, use one channel file per condition".
Import the anatomy
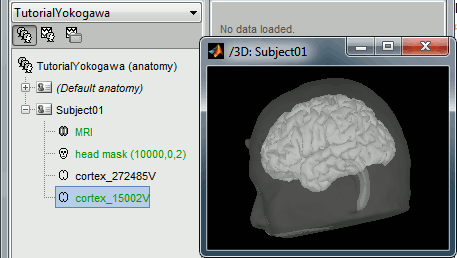
Right-click on the TutorialYokogawa folder > New subject > Subject01
- Leave the default options you set for the protocol
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: sample_yokogawa/anatomy
- Number of vertices of the cortex surface: 15000 (default value)
- Set the 6 required fiducial points (indicated in MRI coordinates):
- NAS: x=128, y=227, z=93
- LPA: x=48, y=130, z=69
- RPA: x=214, y=130, z=76
- AC: x=128, y=148, z=116
- PC: x=128, y=123, z=117
- IH: x=128, y=131, z=176 (anywhere on the midsagittal plane)
At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, which will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.
Without the individual MRI
If you do not have access to an individual MR scan of the subject (or if its quality is too low to be processed with FreeSurfer), but if you have digitized the head shape of the subject using a tracking system, you have an alternative: deform one of the Brainstorm templates (Colin27 or ICBM152) to match the shape of the subject's head.
For more information, read the following tutorial: Warping default anatomy
Access the recordings
Link the recordings
- Switch to the "functional data" view.
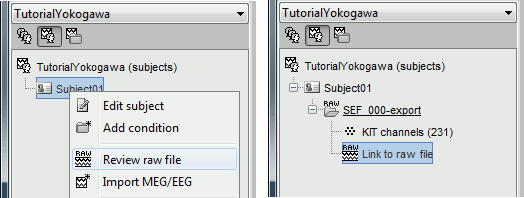
Right-click on the subject folder > Review raw file:
- Select the file format: "MEG/EEG : Yokogawa/KIT"
Select the file: sample_yokogawa/data/SEF_000-export.con
Answer NO when asked to refine the registration using head points. In this dataset, we only have access to the positions of the electrodes and three additional markers on the forehead. The automatic registration doesn't work well in this case, we are going to fix this registration manually.
- A figure is opened to show the current registration MRI/MEG. It is already quite good, but can be improved a bit manually. Close this figure.
- The new file "Link to raw file" lets you access directly the contents of the MEG/EEG recordings
- The channel file "KIT channels" contains the name of the channels and the position of the corresponding sensors.
Prepare the channel file
- The recordings contain signals coming from different types of electrodes:
- 160 MEG channels
- 12 MEG references
- 14 Trigger channels
- 41 EEG channels
- 2 EOG channels: EO1 and EO2
- 1 ECG channel: EKG+
- 1 ground of the EEG amplifier: E
- Not all the types of channels are properly identified in Brainstorm. We need to redefine this manually to get correct groups of sensors.
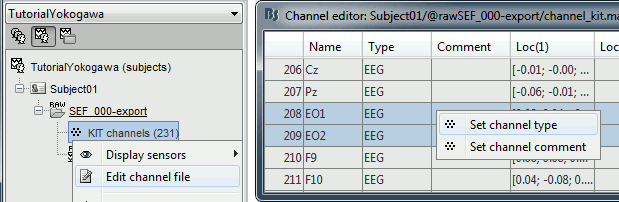
Right-click on the channel file > Edit channel file:
Channel EO1 (208) and EO2 (209): Change the type to EOG
Channel EKG+ (214): Change the type to ECG
Channel E (231): Change the type to MISC
- Close the figure and accept to save the modifications
Refine the MRI registration
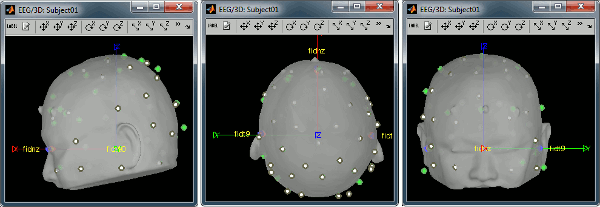
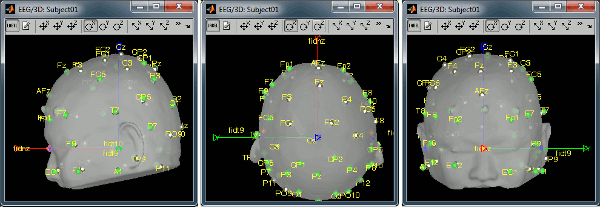
Right-click on the channel file > MRI registration > Edit... (EEG)
- The white points are the electrodes, the green points are the additional digitized head points. To display the label of the electrodes, click on the [LABEL] button in the toolbar. To see what the other buttons in the toolbar are doing and how to use them, leave your mouse over them for a few seconds and read the description.
Now try to manipulate the position of the EEG+MEG sensors using rotations and translations only (no "resize" or individual electrodes adjustments). The objective is to have all the points close to the surface and the three forehead points inside the little peaks on the surface (due to markers in the MRI).
The rotation+translation are going to be applied both to the EEG and the MEG sensors. After you are done with this solid registration part, you can click on the button "Project electrodes on scalp surface", it will help for the source modeling later. The green points (digitized) stay in place, the white points (electrodes) are now projected on the skin of the subject.
- If you feel like you made it wrong, close the figure and cancel the modifications, then try again. It takes a few trials to get used to this rotation/translation interface.
Click on [OK] when done.
Answer YES to save the modifications.
Answer YES again to apply the solid transformation (rotation+translation) to the MEG sensors.
Before manual registration:
After manual registration:
Read the stimulation information
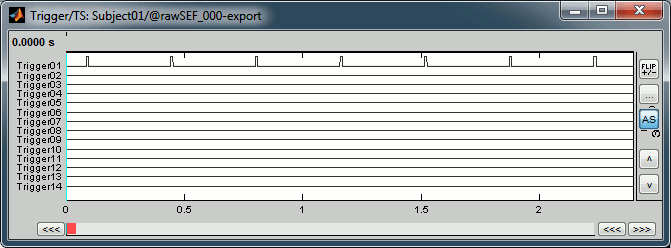
Right-click on the "Link to raw file" > Trigger > Display time series
- In the Record tab, switch to a column view of the sensors (first button in the toolbar)
You can see that all the trigger lines are flat except for "Trigger01", which contains the information of the electric stimulation. We are going to read this trigger channel and convert it to a list of events. Close this figure.
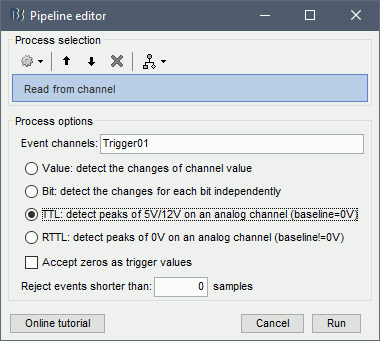
- Drag and drop the "Link to raw file" in the Process1 box.
Run the process "Import recordings > Read from channel"
Event channel = Trigger01
Option selected "TTL": detect peaks of 5V/12V on an analog channel
Do not select the option Accept zeros as trigger values
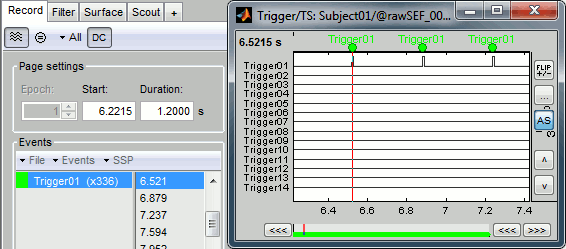
Right-click on the "Link to raw file" > Trigger > Display time series. Check that the peaks of the triggers channel have been correctly identified, then close this figure.
Artifacts: Evaluate the power spectrum
Evaluation
Two of the typical pre-processing steps consist in getting rid of the contamination due to the power lines (50 Hz or 60Hz) and of the frequencies we are not interested in (a low-pass filter to remove the high-frequencies and a high-pass filter to remove the very slow components of the signals). Let's start with the spectral evaluation of this file.
Drag the "Link to raw file" to the Process1 box and run the process "Frequency > Power spectrum density (Welch)". Configure it as illustrated in the following figure:
Time window: [0,50] s, Window length: 4s, Overlap=50%.
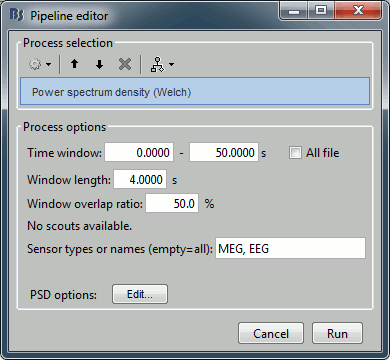
Double-click on the new PSD file to display it.
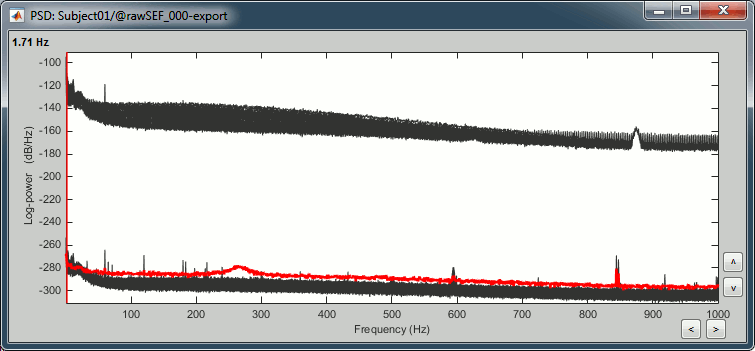
- The lines on the top represent the EEG electrodes, the lines at the bottom the MEG sensors. If you want to get clearer plots, you can calculate separately the spectrum for the two types of sensors separately, by running twice the process "Power spectrum density" , once with sensor types = "MEG" and once with "EEG", instead of running in on both at the same time like we did.
- Observations (below 250Hz):
- Peak around 11Hz: Alpha waves from the subject's brain
Peaks at 60Hz, 120Hz, 180Hz, 240Hz on EEG + MEG:
Electric contamination due to the power lines (60Hz+harmonics)Peaks at 35Hz, 65Hz, 70Hz, 183Hz, 197Hz on MEG only:
Electric noise coming from an unknown sourceMEG sensor LC11 (in red) appears to have a higher level of noise than all the other MEG sensors, we will check this when review the MEG recordings and probably tag it as a bad channel.
Correction
- From the Yokogawa software:
- Users can apply digital filtering to the data before exporting them to Brainstorm. Depending on the data size and degrees of filter parameters, it sometime takes a very long time.
- From Brainstorm:
There is no easy way to process the Yokogawa files at this time using the Brainstorm software. This capability might be added in the future, in which case the procedure would be very similar to the one presented in the basic tutorial ?Detect and remove artifacts.
For short recordings, if the contamination is really bad, there is an alternative option: importing the entire file in Brainstorm and then process it. This approach is illustrated in the EEG/Epilepsy tutorial. This approach requires a very large amount of memory, it is not recommended on MEG recordings as the files tend to be much bigger than EEG-only recordings.
- For now we are going to keep the file as it is. There is no important contamination below 60Hz and we are going to study processes that occur at lower frequencies. When studying evoked responses, the electric noise at 60Hz tends to cancel out and is usually not a problem in the analysis. Note for later that it is however important to consider this electric noise when working on resting state recordings or time-frequency decompositions at higher frequencies.
Artifacts: Heartbeats and eye blinks
Detect artifacts and calculate SSP
Select the "Link to raw file" in the Process1 tab, then create the following analysis pipeline:
Process "Events > Detect heartbeat" on channel EKG+
Process "Events > Detect eye blinks" on channel EO2 (the signal is better than EOG1)
Process "Events > Remove simultaneous"
=> Remove "cardiac" events that are less than 250ms away from a "blink" event.Process "Artifacts > SSP: Heartbeat" for MEG
Process "Artifacts > SSP: Heartbeat" for EEG
Process "Artifacts > SSP: Eye blinks" for MEG
Process "Artifacts > SSP: Eye blinks" for EEG
Review the results
- Double-click on the "Link to raw file" to show the MEG sensors
- By default, only the components with a singular value superior to 12% are selected. This is an empirical threshold that does not work all the time. You should always inspect manually all the spatial components you remove from your recordings, to avoid removing any of the data of interest. We are now going to edit the default selections for those four categories.
Cardiac: EEG: Select component #1 (by default, component #2 was also selected)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Blink: MEG: Select category and select component #1 (by default, nothing was selected)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Blink: EEG: Select component #1 (by default, component #2 was also selected)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
- Click on [Save] to keep your modifications.
Effect on the artifacts
- If we epoch the recordings based on the blink and cardiac artifacts and average the results, we can evaluate what was the influence of the heartbeats and eye blinks and how much of the artifact is removed for each modality with the SSP projections.
You do not need to do this now, but it is an interesting way of studying these artifacts.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Review the recordings
MEG and montages
Right-click on the "Link to raw file" > MEG > Display time series.
- Display the channels in columns (first button in the toolbar of the Record tab).
- Check the list of montages available for this file: click on the drop-down menu in the Record tab.
Select the montage "KIT LC"
Load KIT montages
If you don't see all the "KIT..." entries in this list, load them manually:
Click on All > Edit montages...
- Click on the [Load montage] button
- Go to the folder: brainstorm3/toolbox/sensors/private
- Select the file format "MNE selection files (*.sel)"
Select the file mne_montage_yokogawa.sel
Click on save to close the montage editor.
Bad channels (MEG)
Click on the noisy LC11 sensor to select it (displayed in red)
Press the delete key or right-click in the figure > Channels > Mark selected as bad.
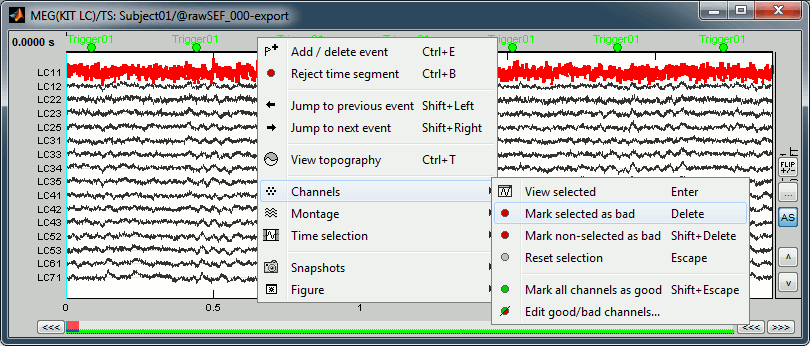
- Close this figure
EEG
Right-click on the "Link to raw file" > EEG > Display time series.
Check the list of available EEG montages.
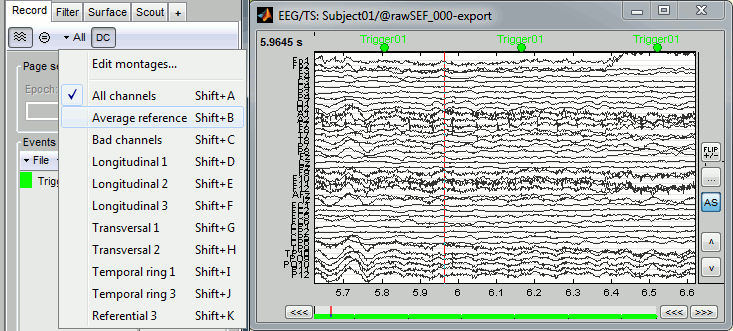
Bad channels (EEG)
In the Record tab, increase the length of the displayed time window to 10s.
You will see that channel TP9 is behaving in a strange way. Select it and mark it as bad.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
More information
We are not going to detail more in details the interface to review MEG or EEG recordings as it is already done in other specialized tutorials:
For continuous MEG: ?Review continuous recordings and edit markers
For continuous EEG: EEG and epilepsy
For imported files: ?Exploring the recordings
Epoching and averaging
Import recordings
In this experiment, the electric stimulation is sent with a frequency of 2.8Hz, meaning that the inter-stimulus interval is 357ms. We are going to import epochs of 300ms around the stimulation events.
Right-click on the Link to raw file > Import in database:
- Select the entire time definition (0s to 120s, default)
Check "Use events" and select "Trigger01"
Epoch time: [-50, 250] ms
Check "Apply SSP" (make sure that it shows 4 active categories / 4 total projectors)
Check "Remove DC offset" > Time range > [-50, -10] ms
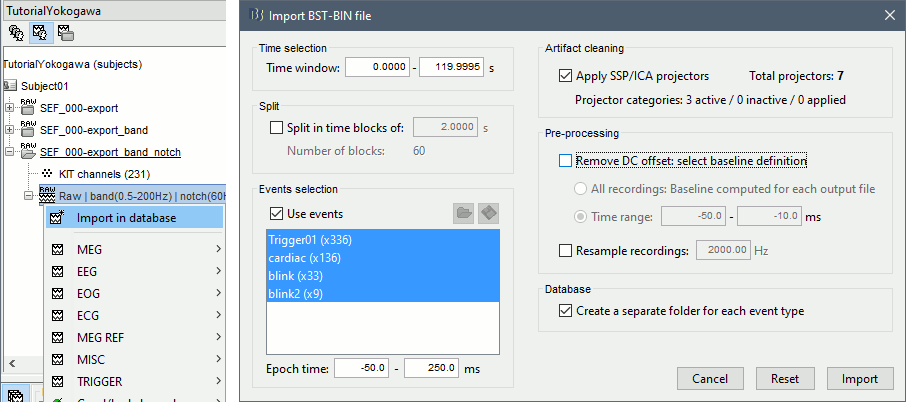
At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 250ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-50,250]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment, we can just ignore it.
Answer Yes to this question to discard the last epoch.
Average epochs
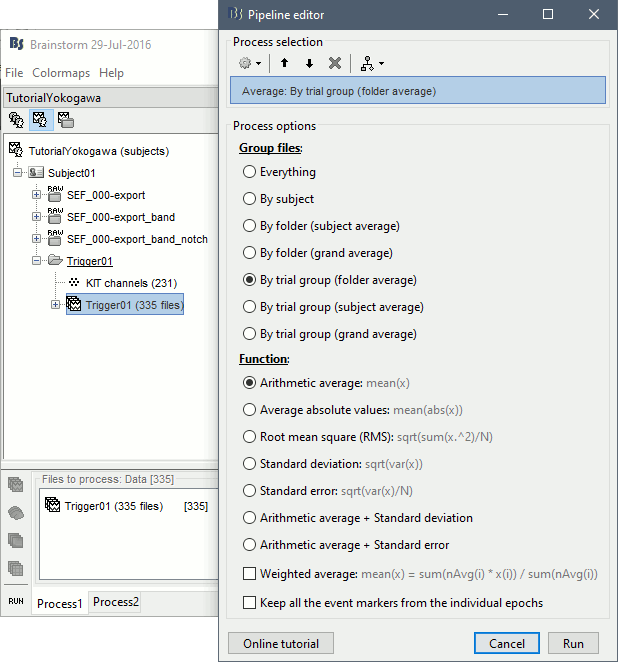
- Drag and drop all the Trigger01 trials to the Process1 tab.
Run the process "Average > Average files".
Review the average for the MEG and the EEG.
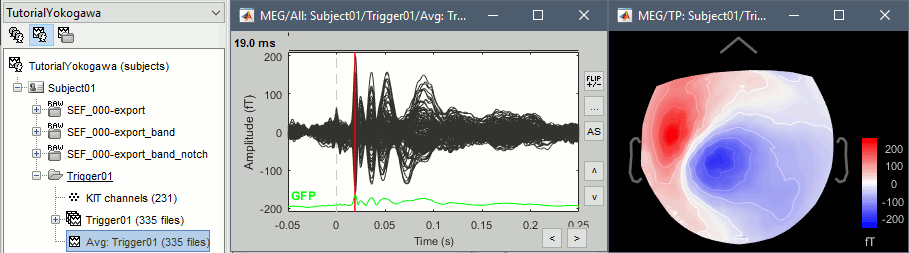
Source analysis
Head model
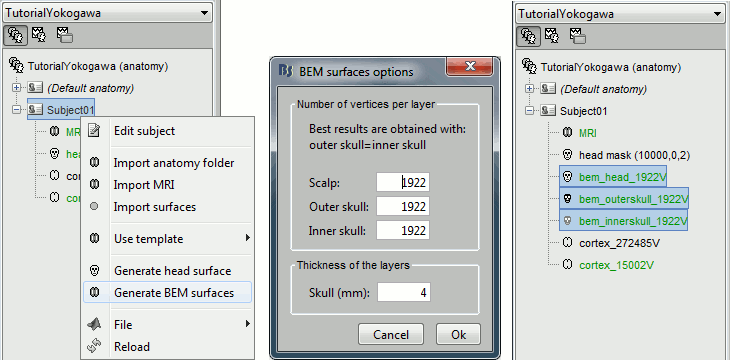
We are going to use a realistic head model, this requires to calculate some additional surfaces for the subject, to represent the inner skull and outer skull surface. Go to the "Anatomy" view, right-click on the subject > Generate BEM surfaces.
Use 1922 vertices for each layer (default).
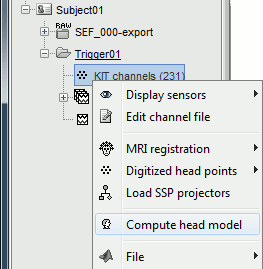
Go back to the "Functional data" view, right-click on the channel file > Compute head model.
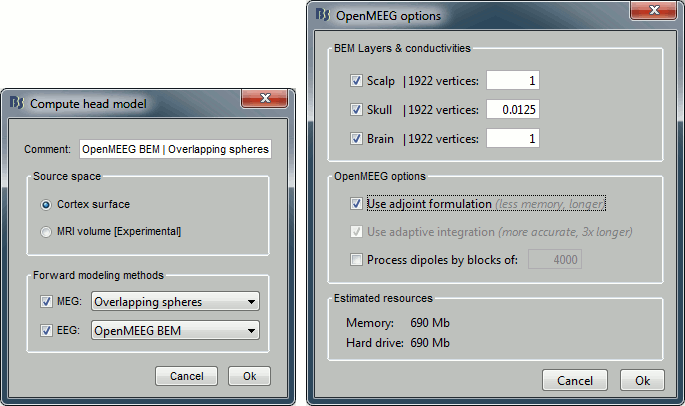
Leave all the default options for the head model (cortex surface, MEG=Overlapping, EEG=OpenMEEG). Then leave all the OpenMEEG options to their defaults except for one: select the option "Use adjoint formulation".
- If you can't download OpenMEEG automatically, download and install it manually (menu Help).
If the OpenMEEG calculation crashes, please refer to the OpenMEEG tutorial.
If you cannot get OpenMEEG to work, or if the results definitely do not make sense, try using a different forward model: "3-shell sphere". It's a spherical model, so it would perform better in the regions of the head that are close to the sphere. See the ?Head model tutorial.
Noise covariance matrix
- We will use the baseline of the single epochs to calculate the noise covariance matrix.
Right-click on the Trigger01 epochs group > Noise covariance > Compute from recordings.
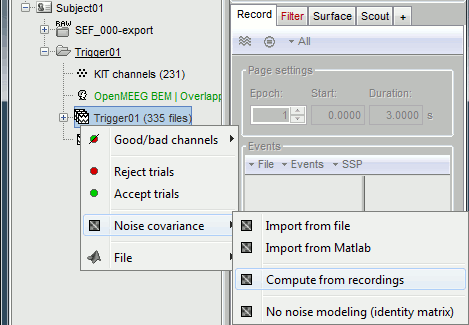
Enter the same baseline interval we used for removing the DC offset: [-50, -10] ms
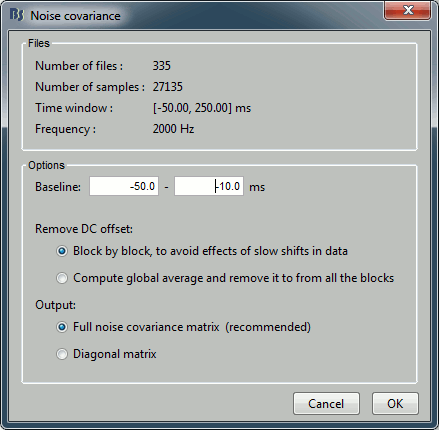
Inverse model
Right-click on the head model > Compute sources.
Select wMNE and MEG. When asked, leave the default list of bad channels (31: channel LC11)
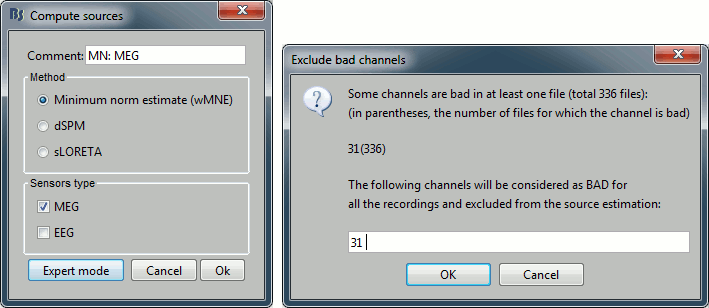
Repeat the same operation for EEG. It is better to study separately the two modalities because the method for combining MEG and EEG are not working well yet.
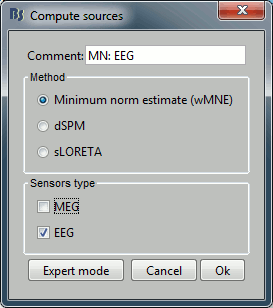
This operation creates a shared inversion kernel and one source link for each block of recordings in the folder. If you are not familiar with those concepts, please refer to the ?Source estimation tutorial.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Display the sources for the MEG (top) and the EEG (bottom).
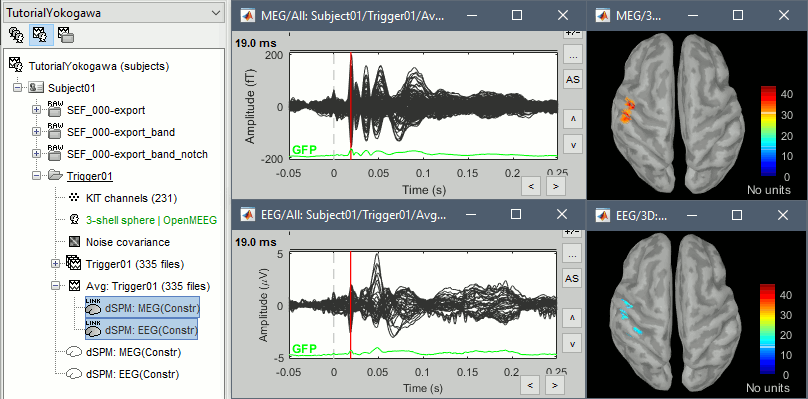
Z-score normalization
- A good way to reveal better the source activity at the cortex level is to calculate a Z-score of the source maps with respect with a quiet baseline. We can use the same baseline as for the calculation of the noise covariance matrix.
- Drag and drop both MEG and EEG average sources in Process1
Run the process: Standardize > Z-score (dynamic), with the baseline [-50,-10]ms
Regions of interest
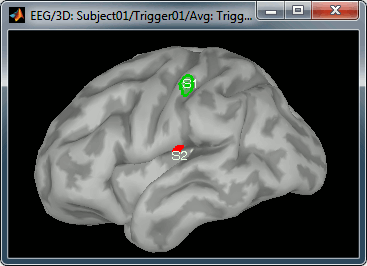
Create two scouts S1 and S2 to represent the primary and secondary somatosensory cortex of the left hemisphere. Proceed with the method described in the contiunous recordings tutorial:
- Open an average source file (eg. MEG Z-score) and the corresponding recordings
- Go to 20ms, adapt the amplitude threshold and the colormap to see only a focal source
- Create a scout at the center of the activated region
- Grow the scout to about 20 vertices and rename it to "S1"
- Go to 50ms and repeat the same operation for S2. In this specific case, S2 does not appear a clear independent region, as we still see stronger activities in more superior regions of the brain. So pick one activated region around the anatomical location of the secondary somatosensory cortex (at the very bottom of the post-central gyrus). See the picture below.
For more information on the scouts, please refer to the ?scouts tutorial.
Then plot the activity for the different files we calculated.
Scripting
The operations described in this tutorial can be reproduced from a Matlab script, available in the Brainstorm distribution: brainstorm3/toolbox/script/tutorial_yokogawa.m
Feedback