Bonjour François,
Thank you very much for your reply.
Regarding the Matlab crash. It is a low-level graphics error, apparently related to Java. Matlab suggested running using OpenGL. I tried this but still cannot open files with "RAW" over them (i.e. links to RAW files) as doing so causes Matlab to crash. However, if I import those same files into the database and opened the imported versions, Matlab does not crash. Files on other data bases that never used to crash are also crashing too. So perhaps something changed on my system relating to Java. I will reinstall Matlab and see if the problem is resolved.
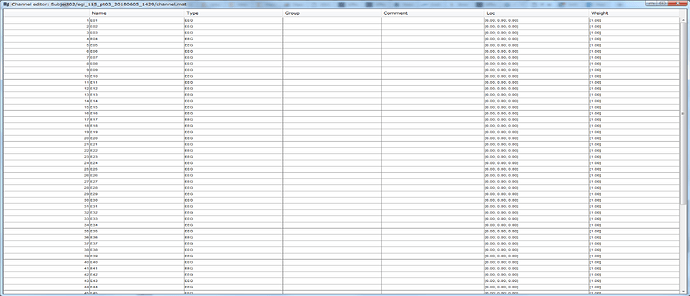
However, the issue with adding EEG channels still remains. The files I am using used a 65 channel setup with ActiCap (I think 64 EEG channels plus one ground channel). However, no matter which template I try and load, I get a message saying that "No channel matching the loaded cap".
Of course, without channel positions, I cannot view sensor topography, and get this error:
BST> Warning: The position of the following sensors is not set: E01 E02 E03 E04 E05 E06 E07 E08 E09 E10 E11 E12 E13 E14 E15 E16 E17 E18 E19 E20 E21 E22 E23 E24 E25 E26 E27 E28 E29 E30 E31 E32 E33 E34 E35 E36 E37 E38 E39 E40 E41 E42 E43 E44 E45 E46 E47 E48 E49 E50 E51 E52 E53 E54 E55 E56 E57 E58 E59 E60 E61 E62 E63 E64 E65
Error using bst_figures>CreateFigure (line 220)
Nothing to display: All the "EEG" channels are marked as bad or do not have 3D positions.
Error in bst_figures (line 59)
eval(macro_method);
Error in view_topography (line 256)
[hFig, iFig, isNewFig] = bst_figures('CreateFigure', iDS, FigureId, CreateMode, DataFile);
Error in bst_figures>ViewTopography (line 1539)
view_topography(DataFile, Modalities{i}, '2DSensorCap', [], UseSmoothing);
Error in bst_figures (line 59)
eval(macro_method);
Error in figure_timeseries>@(h,ev)bst_figures('ViewTopography',hFig,1) (line 2027)
jItem = gui_component('MenuItem', jPopup, [], 'View topography', IconLoader.ICON_TOPOGRAPHY, [],
@(h,ev)bst_figures('ViewTopography', hFig, 1));
The EEG channels are not marked as bad, they just don't have any position information in the channel file.
Do you have any recommendations?
Thank you!
Jared