Dipole scanning and importing
Authors: Elizabeth Bock, Francois Tadel, John C Mosher
In Brainstorm, the brain activity can be estimated with distributed source models including thousands of dipoles and represented as cortical maps or functional overlays on the MRI. Another way of representing the source activity is to consider that at one time point there is only one single dipole that is active and that can explain most of the MEG or EEG recordings.
We offer two options to estimate the position and orientation of the most significant dipole at each time point: a dipole scanning method, which selects the most significant dipole in a grid of dipoles that are already estimated, and a non-linear dipole fitting method, which searches iteratively for the dipole that explains the best the recordings. The former is described on this page, the latter in FieldTrip dipole fitting.
Contents
Dipole modeling
The example below uses the protocol TutorialIntroduction created in the introduction tutorials.
Select the protocol TutorialIntroduction. Go to the view "Functional data (sorted by subjects)".
In the imported Run01, right-click on the head model > Compute sources 2016 > Dipole modeling
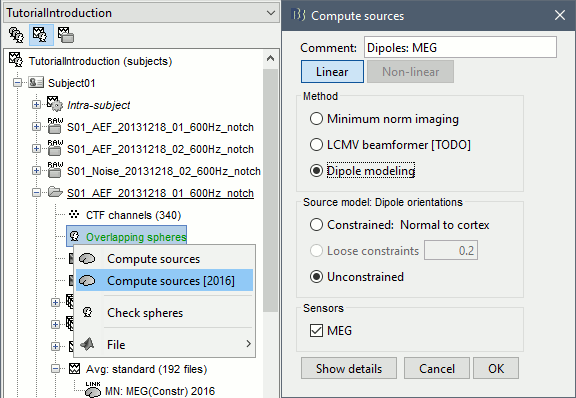
This process allows for a systematic scanning of sources using performance measures to display single dipoles. For this tutorial we will use the Generalized Least Squares (GLS) source modeling available in Brainstorm.
Select the protocol TutorialCTF. Right-click on Subject01 / Left / ERF > Compute sources.
In this window, you should select Expert Mode. You will see the following menu. Select [Test] Mosher GLS. Click OK. Use the default settings in the next menu and click OK.
Two results were added under the ERF: GLS and GLS-P.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
GLS:
GLS-P:
Dipole Scanning
For this tutorial we will do the scanning on the GLS results. However, the dipole scanning is also available for sources computed with minimum norm Z-scored (i.e. MN MEG(Constr) | zscored), dSPM and sLORETA.
Drag and drop the GLS-P link into the Process 1 box. Click Run > Sources > Dipole scanning.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png) ---
--- ![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
- Use the defaults options:
The resulting dipole file is added under the Left condition.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Displaying Dipoles
Right-click ERF|GLSP-dipole-scan > Display on cortex. The Dipoles panel is now open and dipoles are displayed on the cortex. You can now explore the results using the options in the Dipoles panel:
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
- Data options: filter the available dipoles according the specific measurement. For the GLSP, these are defined as:
Z-score:
Goodness:
Chi-square:
Reduced Chi-square:
Confvolume:
- Display options: these options allow for adjusting how the selected dipoles (according to the Data options) are displayed
Show max goodness: will show the dipole with the maximum goodness (within the constraints of the Data options) at the time point selected. If Show all time is selected, then this represents the maximum goodness over all time points.
Show all time: plots all selected dipoles at all time points. Unselecting this options will show only the current time point.
3D - Color dipoles according to Time, Goodness of Fit, Z-score, Group: adjusts the colormap to reflect the measure.
- Data options: filter the available dipoles according the specific measurement. For the GLSP, these are defined as:
In the Dipoles panel, select Show max goodness of fit, Show all time, and color the dipoles according to Goodness of Fit. You should see the following display. Explore the panel options to familiarize yourself with the capabilities.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Additional Example
Dipole scanning can also be restricted to a scout. In this example, the scanning is restricted to a scout named '2' and colored according to Goodness of Fit
Neuromag Xfit and CTF DipoleFit
Brainstorm can import dipoles that have been generated using external software:
Neuromag Source Modeling module, xfit: The .bdip format is described in bdip_format.pdf. For more information about xfit and the bdip format see Neuromag Software Manuals.
CTF DipoleFit: The DipoleFit software and the .dip format is described in the CTF Software Manuals.
To import
- Select the study where you want to import the dipole file
- Right click and select ‘Import dipoles’ (Figure 1)
For Neuromag, you will be prompted to specify the number of subsets used for dipole fitting. In most cases, this will be one. If subsets are used, as defined in the text file $HOME/.meg_analysis/xfit_selections, then the number of subsets used needs to be input when prompted. (Figure 2)
![]()
Figure 1. Importing dipoles
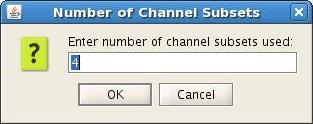
Figure 2. Neuromag option for channel subsets
When displaying dipole files with multiple groups and/or subsets, the Loaded dipoles panel appears with a list of dipoles, grouped according to event (Group #1, Group#2, etc) and by channel subset, if used (Subset #1, Subset#2, etc.)
- Select the Group(s) to plot
- Clicking on a subset will select all Groups with that subset. For example, selecting Subset #1 will select Group #1 (1), Group #2 (1), Group #3 (1), etc.
Examples
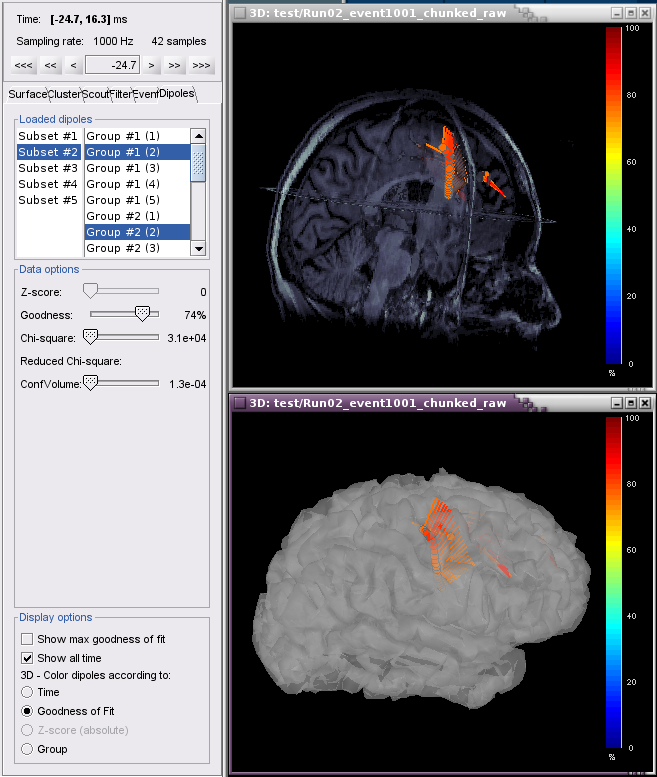
Example 1 shows 5 subsets (channel groups of Left, Right, Anterior, Posterior, All Channels). When subset #2 is selected, dipoles fitted to the right channels are displayed. In this case, Show all time is selected for the 4 Groups (events) and colored according to Goodness of Fit
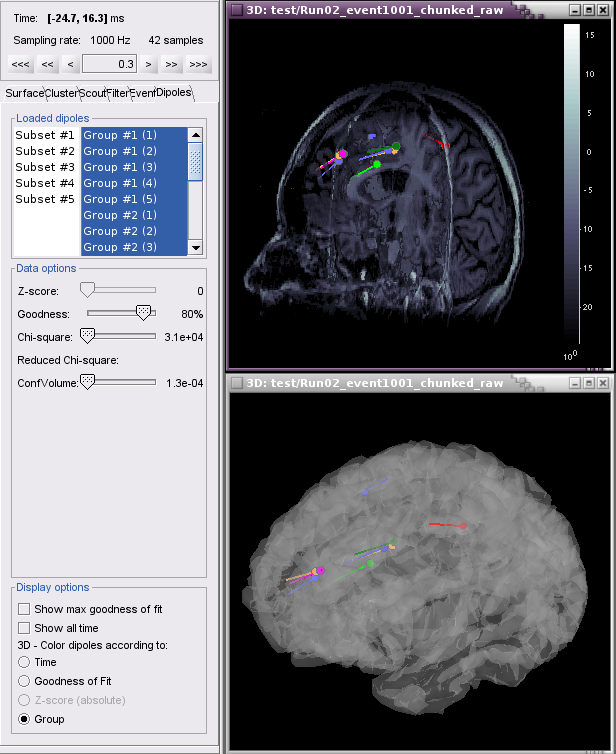
Example 2 shows all groups and subsets at one time point (0.3 ms), with goodness of fit greater than 80%, colored according to Group.
Additional documentation
Tutorial: EEG and epilepsy: Dipole scanning
Tutorial: Dipoles: FieldTrip dipole fitting
Tutorial: Source estimation