Import individual anatomy
Objectives: Import anatomy for CTF MEG tutorial.
Contents
Create protocol
Download bst_tutorial_somatotopy_ctf.zip file (Download section), and unzip it somewhere. Data description in the next tutorial.
Warning: Do NOT unzip it in the Brainstorm database or program directories. This package contains files in original file formats, and need to be imported through the interface to be processed with Brainstorm.
Create a new protocol called TutorialCTF with the following options:
- "No, use individual anatomy", because the individual subject anatomy is available
- "No, use one channel file per condition (MEG)", because we are going to process regular MEG data in the next tutorial
Create a new subject (let's keep the default name: Subject01)
Import MRI
Go to the Anatomy view of the protocol files (very first button in Brainstorm window toolbar).
Right click on subject node, and select menu "Import MRI..."
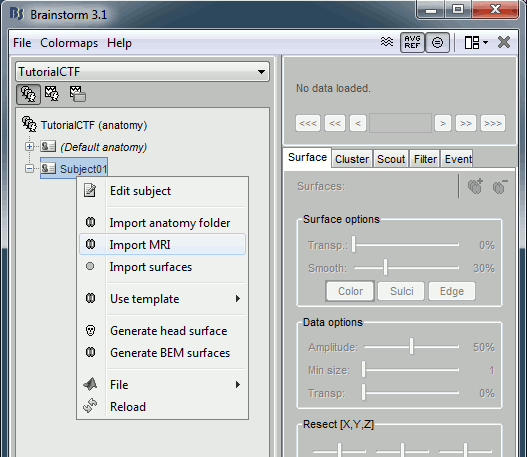
Select file "TutorialCtf/Anatomy/01.mri".
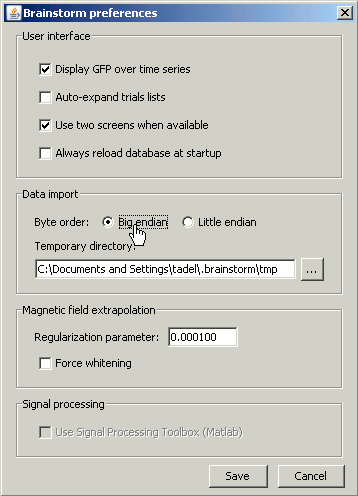
If you didn't modified any Brainstorm option yet, you should get an error message, saying "Unrecognized format, try switching byte order". This is because not all the computers process the binary data the same way. To learn more about endianness, you can consult this Wikipedia page. Depending on the type of computer which wrote the binary files, you may need to read them as little-endian or big-endian. To change this setting, go to the Options > Byte order menu in main window. You may need to modify this setting again to import other files.
Try to import the MRI again. It should be working now. The MRI viewer is displayed, together with a message box that tells you what to do. Follow the instructions. The MRI is already well oriented so you can directly process with the fiducials selection. You will see that three fiducials were already identified in the MRI file (NAS, LPA, RPA); you just need to add the three other ones (AC, PC, IH). If you are not sure how to do this, consult this page: CoordinateSystems.
Import surfaces
The MRI is now available in the anatomy folder for Subject01. Let's now proceed with the surfaces. The envelopes we are going to import in this tutorial were extracted using ?BrainVISA software, and their names and types may differ from other software solutions. The next tutorials will show you how to import surfaces from other programs (?BrainSuite, ?FreeSurfer).
Right click on the subject node again, and click on "Import surfaces...". Select at the same time (holding Shift or Ctrl key) all the files in the directory "TutorialCtf/Anatomy/BrainVISA" (head, Lhemi, Rhemi). Click on "Open" and answer Yes to the question: "Align surfaces with MRI now?".
- Note: The coordinate system used to localize in 3D the vertices of each surface depends on the software that created them. When importing the surfaces, Brainstorm registers them with the MRI using its fiducials points (NAS, LPA, RPA). This simple algorithm needs the head (ie. scalp) surface, so if you try to import only the cortical surface, it will not be well registered with the MRI, and all the following computations will fail.
You should see the three surfaces in the database tree.
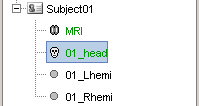
One has been identified automatically as a scalp (=head) surface, because its original filename contained the "scalp" keyword. Usually, Brainstorm identifies file types with this kind of tags in the file names.
The two other files where not identified automatically. They contain the envelopes of left and right hemispheres of the brain, but Lhemi and Rhemi are not keywords that are identified automatically by Brainstorm.
Try to display the surfaces by double-clicking on them, or with the popup menu Display.
Downsample and merge surfaces
In the Surfaces tab, you can see the numbers of faces and vertices for each surface, and also display their faces (wireframe) with the Edges button. They look great, but their resolution is too high for the algorithms we will apply on them later, they need to be downsampled.
- Downsample:
Right-click on the head surface > Less vertices...: enter 7000, and press Ok.
Right-click on the Lhemi surface > Less vertices...: enter 7500, and press Ok.
Right-click on the Rhemi surface > Less vertices...: enter 7500, and press Ok.
- Three new surfaces appeared in the list, with the number of vertices as a suffix.
Group left and right hemisphere to build a Cortex surface:
Select Lhemi_7500V and Rhemi'_7500V' files at once (hold Ctrl key and left-click on both)
Right-click > Merge surfaces
A file called New surface appeared at the end of the list, rename it in Cortex (F2 key, or click twice on it but waiting between the two clicks, or right-click > Rename). Its icon should change automatically: the surface has been classified automatically as a Cortex (keyword that was recognized).
Double-click on it to display it (or right-click > Display), and check in the Surfaces tab that the number of vertices is about 15000.
- A bit of cleaning:
Rename the 01_head_SCS_7000V in Head.
Delete all the surfaces but the Head and Cortex (Delete key, or right-click>Delete).
Before:
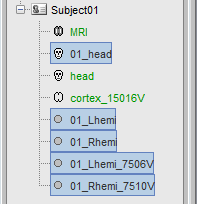 After:
After: 
Bug warning
Description: On some operating systems, it may happen that the tree is not refreshed well. Just after adding new files, the database tree can be displayed incompletely or can event appear completely empty. This bug occurs sometimes because the interactions between Matlab and Java are not always well synchronized.
Workaround: If this happens, just refresh the tree display with by either pressing the F5 key, or by click on the menu File > Refresh tree display. We'll try to fix this bug some day.
Check registration with MRI
Your Subject01 anatomy is ready for source estimation. But before going further, you should always check that surfaces and MRI are well registered. When performing those steps a bit too fast, it is really easy to end with a cortex envelope that is not aligned with the MRI. None of the following steps in source estimation will check that for you, and the sources estimated would be completely wrong.
Right-click on the Cortex surface > Align > Check alignment with MRI...
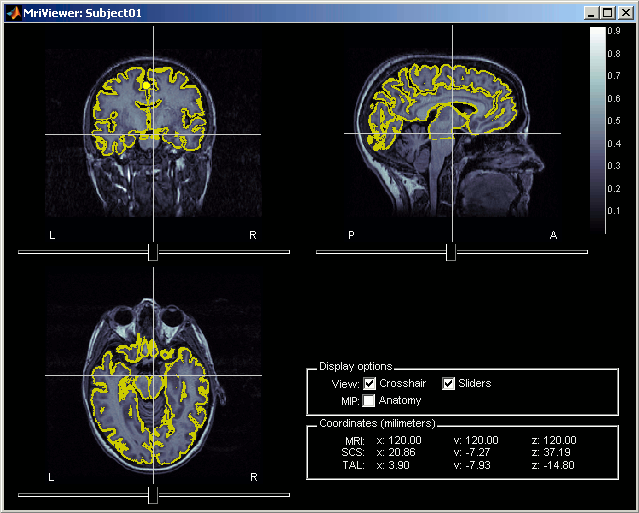
- The yellow lines represent the re-interpolation of the surface in the MRI volume. The computation may take a while, but the result will be stored in the surface file, and will be useful for other operations anyway.
Fix registration with MRI
In some rare cases, the surfaces may not be registered correctly with the MRI. If this happens, delete your MRI and surfaces and start again from scratch. If the result is not better and you don't know what happens, there are still two emergency options:
Defining the fiducials on the head surface. Of course it is possible only if you have the head surface. Right-click on the surface > Advanced > Align all surfaces (with MRI fiducials)... You will be asked to point on the scalp surface the same fiducials as in the MRI (NAS, LPA, RPA).
- the manual alignment...
What happend on the harddrive
If you are curious to know how all this anatomy information was stored in the database directory on the hard drive, you can have a look in your brainstorm_database/TutorialCTF directory.