Tutorial 3: Importing MEG recordings
We are now going to import the recordings you downloaded in the previous tutorial (bst_sample_ctf): the averaged reponses to electric stimulation of the thumbs on both hands, 400 trials for each hand. Stimulus occurs at time 0.
Contents
Import recordings
Select the TutorialCTF protocol, and go to the Functional data (sorted by subject) view of the database.
Right-click on Subject01, select Import MEG/EEG...
File format: "MEG/EEG: CTF .ds (*.meg4;*.res4)"
File: bst_sample_ctf/data/somMDYO-18av.ds (select the the full directory and click on Open)
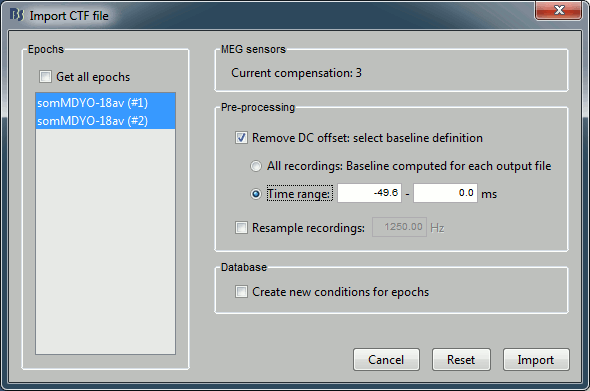
- Two blocks of data are available in this file. The names are not really meaningful, so you have to know what was saved in the file. In this specific case, the first "epoch" is the average, the second is the standard deviation for the same trials. Keep both "epochs" selected.
Check the "Remove DC offset" option, together with "Time range", between -49.6 and 0. For each sensor, this will compute the average value across the time (on the for the pre-trigger period, ie. where times < 0), and substract this mean to all the time values.
=> In MEG, this operation is always needed, unless it was already applied during one of the pre-processing steps.- Click on Import.
A condition was created, called after the filename. It contains three items:
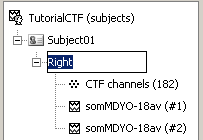
The first is the channel file: description of the positions, names, types, and various properties of the sensors that recorded the data. The value (182) means that there are 182 channels of data in the data files. They are not necessarily MEG channels, it may also includes EEG, EOG, stimulation lines, references, etc.
- The last two represent the recordings associated with the stimulation of the Right thumb (average and standard deviation).
- Having the channels defined in the same file as the recordings is specific to MEG. Those systems always store all the available information at the same time. When dealing with EEG, you have to import separately the positions of the electrodes. This will be developped in another tutorial.
Rename the condition into Right (F2, or successive left clicks, or right-click>Rename)
Display channel file
Let's explore what you can do with the first file. Right-click on the CTF channels file and try all the menus.
Menu: Display
The two menus in the Display menu display the same thing, but in a different way. You can add the scalp surface easily with the toolbar in the Surfaces tab, in the main window (Add a surface button).
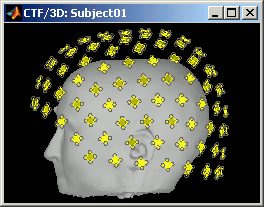
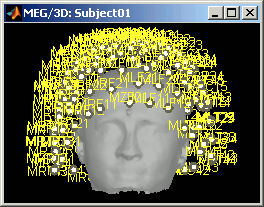
CTF axial gradiometers (only available for CTF recordings):
- All the CTF MEG axial gradiometers (only the coil close to the head is represented).
- This menu displays them as they will be used in the forward model computation. The small squares do not represent exactly the reality, as CTF coils are circular, but they are modelized like that.
MEG: MEG sensors are now represented as small white dots (center of the coils close to the head), and tesselated.
Menu: Edit channel file
Display a table with all the information about the individual channels. You can use this window to view and edit the channels properties.
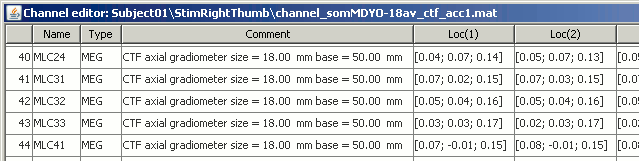
The channel file describes each channel separately, with the following information:
Indice : Indice of the channel, which is also the indice of the row in the data matrix ('F' field in the recordings file). You can edit this field if you need to reorder the channels (might be useful for EEG recordings).
Name : Name that was given to the channel by the acquisition device.
Type : Channel type, eg. MEG, EEG, EOG, ECG, EMG, Stim, Other, etc.
- Sometimes you have to change the Type for some sensors. For instance if the EOG channel was saved as a regular EEG channel, you have to change its type to prevent it from being used in the source estimation.
Comment : Description of the channel.
Loc : Indicates the position in space of the sensor (x,y,z coordinates). One column per coil and per integration point. You should not modify these values from this interface.
Orient : Indicates the orientation of the coil (x,y,z coordinates). One column per coil or per integration point.
Weight : When there are more than one coil or integration point, the Weight field indicates the multiplication factor to apply to each of these points.
Menu: File
Some other fields are present in the channel file that cannot be accessed with the Channel editor window. You can explore those other fields with the File menu, selecting View file contents or Export to Matlab. As we saw in previous tutorial.
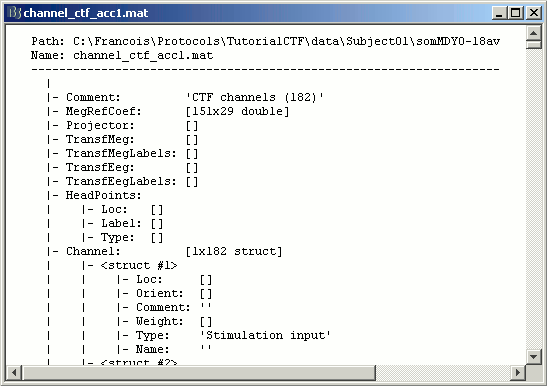
Some fields you may find there:
Comment : String that is displayed in the Brainstorm database explorer
MegRefCoef: Noise compensation matrix for CTF recordings, which is computed by the CTF systems, based on some other sensors that are located far away from the head.
Projector: SSP projectors used for denoising purposes.
TransfMeg: Transformations that were applied to the positions of the MEG sensors to bring them in the Brainstorm coordinate system (as Brainstorm and CTF coordinate systems are the same, this field is empty).
TransfEeg: Same for the position of the EEG electrodes.
HeadPoints: Extra head points that were digitized with a tracking system (eg. Polhemus Isotrak).
Menu: MRI Registration
For the moment, the registration between anatomy and sensors is based only on three points that are manually positioned (nasion and ears). This rough alignment technique is quite robust but also very unprecise, and depends on the precision with which the people defined the fiducials, both during the data acquisition and on the MRI slices.For this reason, it is sometimes necessary to correct the position of the sensors. Two options:
Automatic Registration
Menu MRI Registration > Auto registration (using head points).
If some digitized head points are available, this menu launches an iterative algorithm that uses those points to try to get a better MRI/sensors registration. Here, there are no such points that were acquired, and you will get an error message if you select this menu.
Manual registration
From a scientific point of view it is not a really rigorous operation, but sometimes it is much better than using wrong default positions.
There are two menus: one to check the alignment head surface/sensors, and one to fix it. The Manual MEG-MRI registration menu works exactly the same way as the surface alignment tools introduced in previous tutorial (select buttons in the toolbar and right-click+move in the windows).
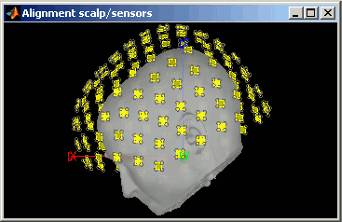
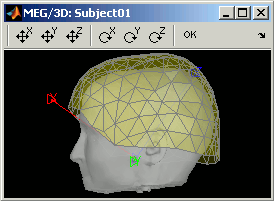
Nothing to change here, but remember to always check the registration scalp/sensors just after you import MEG or EEG recordings.
Display recordings
To understand what is stored in the two recording files: double-click on the first, then double-click on the second.
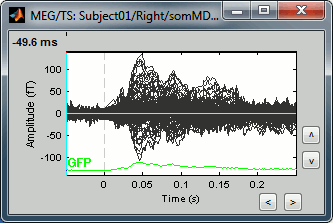
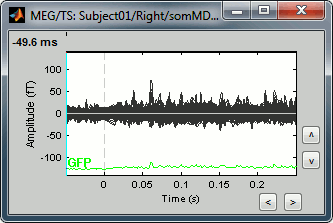
- The first one is the response to the electric stimulation averaged over 400 trials across the time (the values of each MEG sensors is represented by a black line, and they are all overlaid). Roughly, you can observe a first response peak at 23ms, and a second one at about 47ms.
- The second one is the standard deviation of the trials that were average, for each sensors and at each time.
You can rename them ERP and Std (or Error), because it makes more sense than somMDYO-18av (#1) and somMDYO-18av (#2).
Close the windows using the Close all button on the right of the main Brainstorm toolbar. It closes all the figures at the same time and frees all the memory. Try to always use this instead of closing the figures individually.
Now have a look to what is inside a recording file: right-click on Right/ERP > File > View file contents.
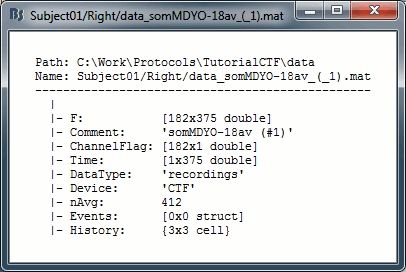
F: recordings values (nChannels x nTime), in Volts
ChannelFlag: one value per channel, 1 means good channel, -1 means bad channel (and will not be displayed or processed)
Time: Time values, in second
Device: Name of the acquisition system used to record this file
nAvg: For averaged files, number of trials that were used to compute this file
History: Operations performed on file since it was imported
DataType: Type of the data saved in the F file (recordings, zscore...)
BadTrial: 0 if this block of data is accepted as a valid data file, 1 if the file was classified as unusable (automatically or manually)
Comment: String displayed in the Brainstorm database explorer to represent this file
All the recordings display tools will be introduced in the next tutorial.
Managing conditions
You have already imported the average response for the stimulation of the right thumb. Let's also import the left thumb, but proceeding in a slightly different way.
Create a new condition for your Subject01, called Left. To do that: right-click on Subject01 > Add condition...
Import the second file (somMGYO-18av.ds) in the Left condition. Rename the recordings in ERP and Std.
Switch to the Functional data (sorted by condition), by click on the third button in main Brainstorm toolbar.
- This view is not very useful when you have one subject and two conditions. But remember this option for later, it will be very useful when you will have 60 subjects and 12 conditions per subject in your database.
Next
Everything is now loaded in TutorialCTF protocol : subject anatomy (MRI and surfaces), recordings and channels description.
The next tutorial will present all the tools available to ?explore the recordings.