Tutorial: Source estimation
This tutorial is still based on Sabine Meunier's somatotopy experiment, called TutorialCTF in your Brainstorm database.
Now you have in you database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the results. Many solutions described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust.
For more information about inverse models and minimum-norm estimation, visit the ?Theory section.
Contents
Computing sources for a single data file
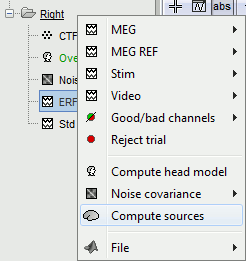
Right-click on Subject01 / StimRightThumb / ERP > Compute sources.
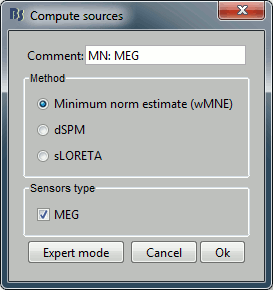
- With this first window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation.
- Edit the Comment field if you want.
- Select "Min. Norm Imaging"
- For the sensors, you have no other option than MEG (All), because those recordings were recorded with only one type of MEG sensors (axial gradiometers) and no EEG signal was registrated at the same time.
- Run.
The next window shows the options for the minimum norm estimation algorithm.
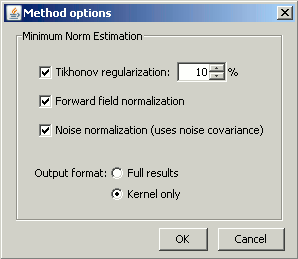
Leave the Tikhonov and Forward field normalization selected. To learn more about those options, please consult the the ?Theory section.
However, you need to understand well what is the meaning of the last option, Output format, which defines the way the sources estimates are saved.
Full results: Saves in one big matrix the values of all the sources (15000) for all the time samples (375).
- The size in memory of such a matrix is about 45Mb for 300ms of recordings. This is still reasonable, so you may use this option in this case.
- But if you need to process longer recordings, you may have some "Out of memory" errors in Matlab, or fill your hard drive quicklky.
Kernel only: Saves only the Inversion kernel, a matrix which describes how to compute the sources when you know the values at the sensors level. So its size is: number of sources (15000) x number of sensors (151).
- To get the sources time series, you just need to multiply this kernel by the MEG recordings.
Full results = Inversion kernel * Recordings
- The size of this matrix is about 18Mb. In this case, the difference is not very important because we only process 375 time samples. But this inversion kernel is independent from the recordings length, so you can easily scale its computation to much longer recordings.
- This is possible because the Minimum norm estimation is a linear method.
- Which option should you choose ?
Probably "Kernel only", as it is faster and produces smaller files.
- All the following operations in Brainstorm will be exactly the same whatever you select. Each time you will access the sources values, the program will have to do the multiplication Kernel * Recordings, but this is done in a totally transparent way.
The only reason that would make you chose the "Full results" options would be any interest in having the full matrix in one file, in case you want to process the sources values by yourself (filtering, statistics, display...).
So please "Kernel only" for the moment. And click Ok.
A new file is available in the database explorer.
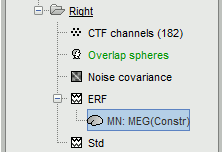
It is displayed inside the recordings file ERP, because it is related to this file only.
You can have a look to what there is in the corresponding matrix file (right-click > File > View .mat file). You would find all the options of forward and inverse modelling, and only one interesting field : ImagingKernel.
Sources visualization
There are two main ways to display the sources: on the cortex surface and on the MRI slices.
Sources on cortex surface
Double-click on recordings StimRightThumb / ERP, to display the time series (always indispensable to have a time reference).
Double-click on sources StimRightThumb / ERP / MN: MEG(Kernel).
Equivalent to right-click > Cortical activations > Display on cortex.Go to the main peak around 45ms (by clicking on the times series figure)
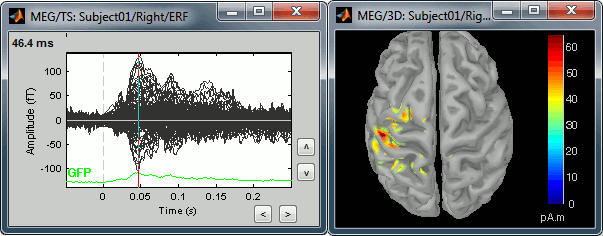
Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recodings figures: rotation, zoom, Surfaces tab(smooth, curvature, resection...), colormap, sensors, pre-defined orientations (keys from 1 to 6)...
Only two new controls are available in the Surfaces tab, in panel Data options: Threshold and transparency.
- Transparency: changes the transparency of the sources on the cortex.
- Threshold: Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed.
Take a few minutes to understand what this threshold value represents.
The colorbar maximum depends on the way you configured your Sources colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 55 pA.m.
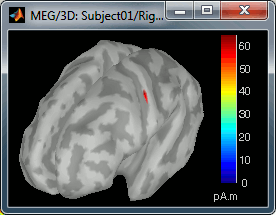
- On the screenshot above, the threshold value was set to 34%. It means that only the sources that had a value over 0.34*55 = 18.7 pA.m were visible.
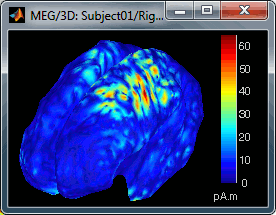
- If you set the threshold to 0%, you display all the sources values on the cortex surface; and as most of the sources have values close to 0, the brain is mainly blue.
Move the slider and look for a threshold value that would give you a really focal source.The following figures represent the sources activations at t=46ms respectively with threshold at 0% and 90%.
Sources on MRI
Close all the figures (Close all button). Open the time series view for StimRightThumb / ERP.
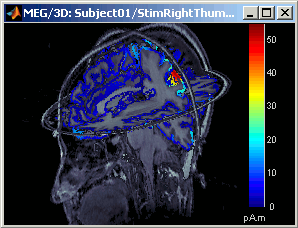
Right-click on StimRightThumb / ERP / MN: MEG(Kernel) > Cortical activations > Display on MRI (3D).
This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.
Modelized data
What is called modelized data here is the part of the recordings that are explained by the minimum-norm solution. This information is saved the sources file, in the Fsynth field.
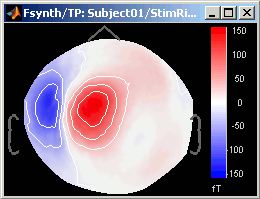
Close all the figures. Open the 2D sensor cap view for StimRightThumb / ERP.
Right-click on ERP sources file > Modelized data > Display time series
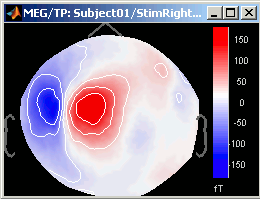
Right-click on ERP sources file > Modelized data > 2D sensor cap
You can browse among time to observe what are the differences between what the model explains and the real recordings. The modelized data is smoother: the regularization which is applied to obtain the sources is supposed to lower the noise.
Residuals
The residuals are the difference between the recordings and the modelized data. This information is not stored in any file but reconstructed when needed.
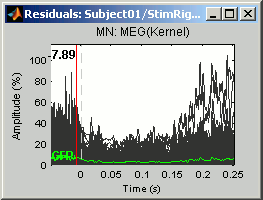
Right-click on ERP sources > Residuals > Display time series
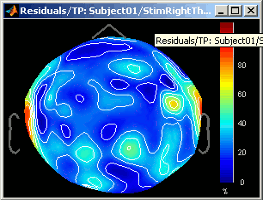
At each time: Residuals = || Modelized - Recordings || / || max(Recordings)Right-click on ERP sources > Residuals > 2D sensor cap: Spatial representation of the same values.
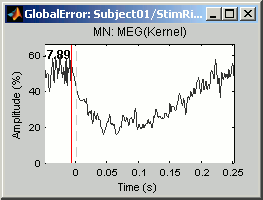
Right-click on ERP sources > Residuals > Display global error:
At each time: Norm of residuals across all the sensors / Norm of the recordings across all the sensors