Thank you for your reply Francois!
So I decided to follow your advice about:
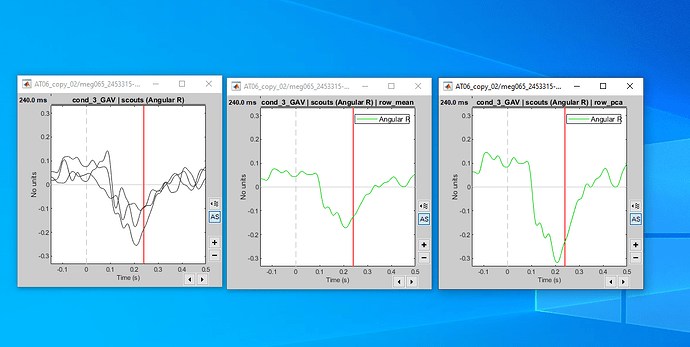
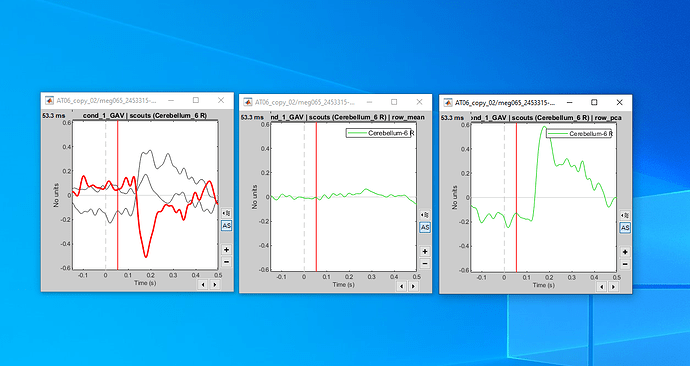
computing the connectivity matrix for all the orientations (x,y,z), and take the maximum connectivity measure across the three orientations.
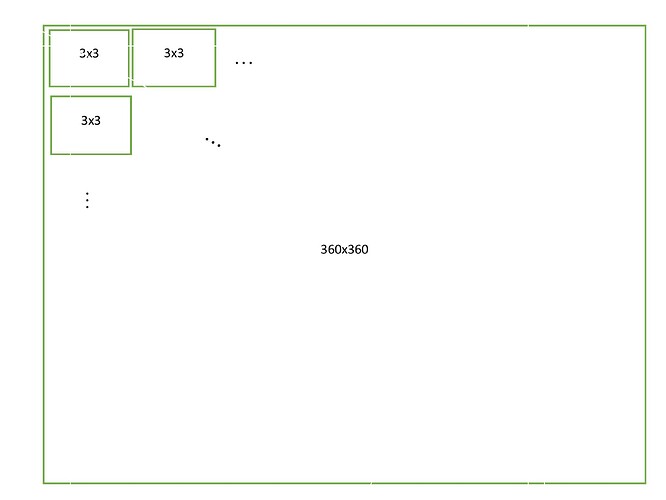
I work with AAL2 parcellation and I have 120 scouts. Because of the three source directions for each scout I get a 360x360 adjacency (connectivity) matrix.
In order to find the maximum connectivity measure for each xyz direction of each scout, I find the maximum value for all the 3x3 matrices inside the 360x360 adjacency matrix:
What do you think of this approach?
I will also upload my matlab script that does the aforementioned. It might be helpful for someone that needs to do a similar reduction in a adjacency matrix:
%% reducing the 3 source directions to 1 source direction per scout (matrix 360x360 becomes 120x120)
%% 3*43=129 adjacency matrices which means 43 subjects under 3 different conditions
%% Method used to find the adjacency matrices: Transfer Entropy (TE)
structure = load('TE.mat') ; %load structure
field=structure.indexes.TE.data; %load field
[rownum1,colnum1]=size(field);
[rownum,colnum]=size(field{1,1});
for i = 1 : rownum1
for j = 1 : colnum1
new_adjacency_matrix=zeros(rownum,colnum);
adjacency_matrix=field{i,j}; % read the one of the 129 adj. matrices
for k1 = 1:3:rownum-2 %1, 1+3=4 , ... , 155+3=158 -> 120 times
for m1 = 1 : colnum
Rows_Comparison = adjacency_matrix(k1:k1+2,m1);
maximum=max(Rows_Comparison); %max(Rows_comparison) is a row vector 1x1 containing the maximum value of each column
new_adjacency_matrix(k1,m1)=maximum; %create 360x360 matrix
end
end
new_adjacency_matrix( ~any(new_adjacency_matrix,2), : ) = []; %delete rows with zeros
%We have now a new_adjacency_matrix: 120x360
for k2 = 1 : rownum/3
for m2 = 1:3:colnum-2 %1, 1+3=4 , ... , 355+3=358 -> 120 times
Columns_Comparison = new_adjacency_matrix(k2,m2:m2+2);
maximum=max(Columns_Comparison,[],2); %max(Columns_comparison) is a column vector 1x1 containing the maximum value of each row
new_adjacency_matrix(k2,m2)=maximum; %create 120x360 matrix
end
end
%Now we equate the columns: 2,3, 5,6, 8,9,... and so on with 0. That way we can get rid of them afterwards
for k3 = 1 : rownum/3
for m3 = 1:3:colnum-2
new_adjacency_matrix(:,m3+1)=0;
new_adjacency_matrix(:,m3+2)=0;
end
end
new_adjacency_matrix( :, ~any(new_adjacency_matrix,1) ) = []; %delete columns with zeros
TE_adjacency_matrix = new_adjacency_matrix- diag(diag(new_adjacency_matrix)); %make diagonal elements equal to 0
save(['TE_adjacency_matrix_' num2str(i) '_' num2str(j) '.mat'],'TE_adjacency_matrix');
%We have now TE_adjacency_matrix: 120x120 with diagonal elements equal to 0
end
end