Hello,
I'm doing source connectivity (1xN coherence) on resting state data that has variable length in different subjects. When choosing "All file" for the time window and "save average connectivity matrix" as the output, BST throws an error if any of the subject files have a time length shorter than the first file. Are there any suggestions for how to achieve a similar output without the time errors?
1 Like
All the methods available need signals of the same duration (exact same number of time samples).
If your recordings have different duration, you need to select a common time window across all the files (and therefore NOT use the option "All file", or import the recordings before)
Thanks, Francois. I know the connectivity tutorial is still in the works, but one more question. As an alternative, would it be appropriate to calculate individual connectivity matrices for each subject (all of different time length) and then just average them after? Is that the same process behind the built in "save average matrix" output?
I'm sorry, I misunderstood your question initially: I thought you wanted to compute a connectivity measure between signals of different duration. But obviously this is not the case.
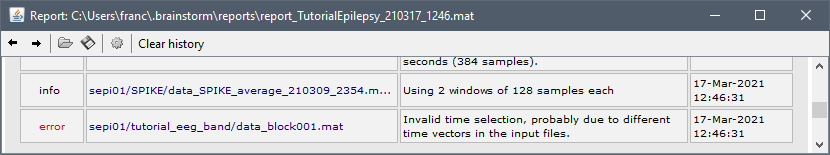
If I understand well, you have multiple recordings of different duration, you put them all in the Process1 box and run the Coherence [1xN] process with the option "Save average connectivity matrix". But Brainstorm does not let you do this and you get an error message like this one:
What you are trying to do is correct for the estimation of Coherence, but the pre-loading of the files by bst_connectivity doesn't let you do it, as not all the connectivity metrics would work in this configuration.
As a workaround, yes you can compute the coherence values separately for each file and average them after. What you can do is to select the option "Save individual results" instead, followed by the Average process. To be tested, but I think it should give you the same results.
Hi thanks! That was exactly my question. All the best!
Hi Francois, in line with this remark: I have resting state EEG data of subjects under two different conditions (pre and post meditation) and I wish to do NXN connectivity analysis (either PLV or Amplitude Envelope Correlation) to see how the resting state connectivity differs between the two conditions of interest.
My only issue is that my EEG pre-processing pipeline is excluding some bad channels without interpolating them. Will it be a problem for statistical analysis if the channel numbers differ across subjects/conditions?
The t-test on connectivity matrices need to have files in input with exactly the same dimensions, therefore yes, it would be a problem to have different lists of channels.
You can put all your imported recordings into the Process1 tab and run the process "Standardize > Uniform list of channels".
/!\ Make a full backup of your database before trying this out, as it could damage seriously your database structure if anything goes wrong.
1 Like
Thanks, I re-ran my pre-processing pipeline to ensure uniform number of channels!