Hi Raymundo, hi all,
I found a way to use the mrk file without head points with some modification of the in_fopen_kit function:
in_fopen_kit_ber.m (28.6 KB)
It looks as if it works, although I don't know the ground truth of course. Do you think this makes sense?
Thanks again,
Bahne
Hi @BahneBahners, apologies for the long delay.
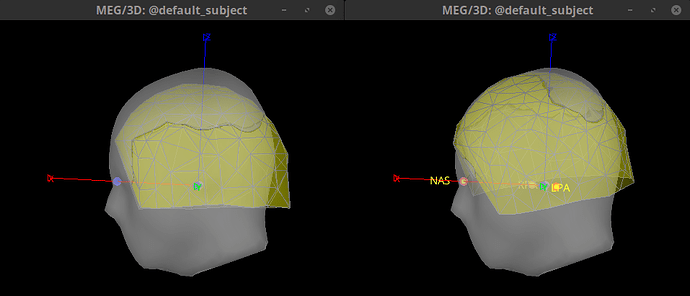
In theory it makes sense to use the position of the position coils as an approximation to the fiducials. However, the locations that are saved in the .mrk file do not help for the coregistration, they are quite random, check the gif below. Axis are made with the anatomical fiducials. Yellow points (with red border) are the positions of the NAS, LPA and RPA coils according to Yokogawa_file.mrk.
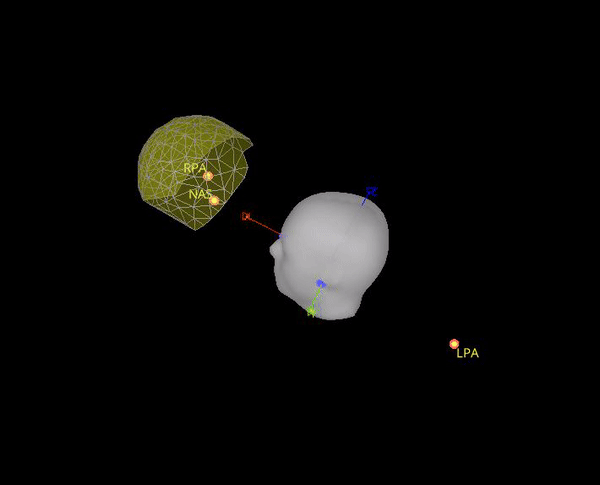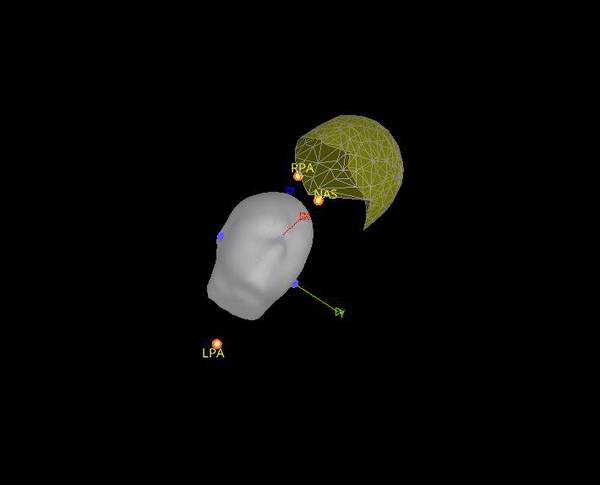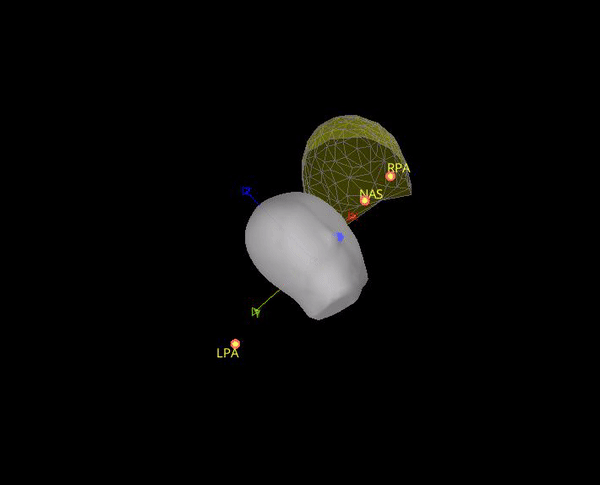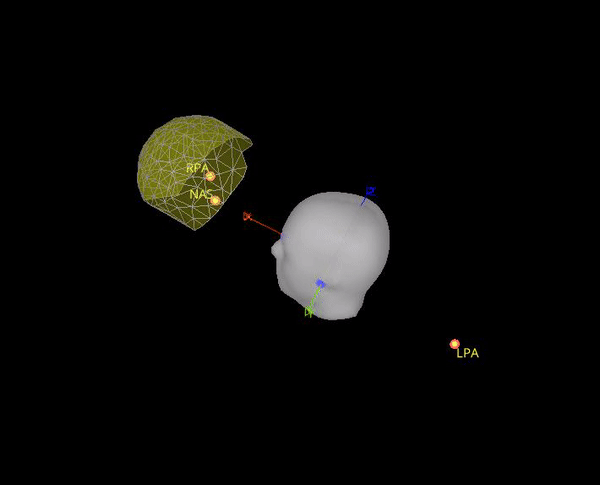
It seems this is note case, the positions for the 5 HPI are different. Moreover, the content of Yokogawa_file.mrk, indicates that it comes from this file: plfp25.before2.mrk , while the Yokogawa_file-coregis.txt points to the file plfp25.before2_bp25-100.mrk.
Do you know something about why there were at some point two marker files?
Or more info about the .mrk file related to Yokogawa_file-coregis.txt?
As the marker files are different, in test below the locations for NAS, LPA and RPA from Yokogawa_file-coregis.txt we used. While the co-registration is better, it is not perfect, it's likely the coils were not placed on the fiducial locations.
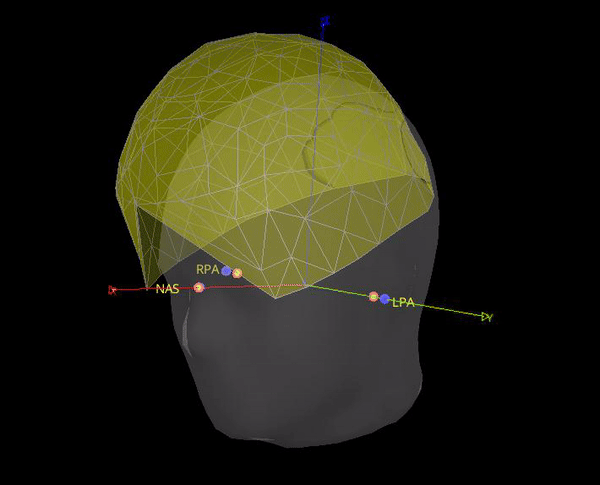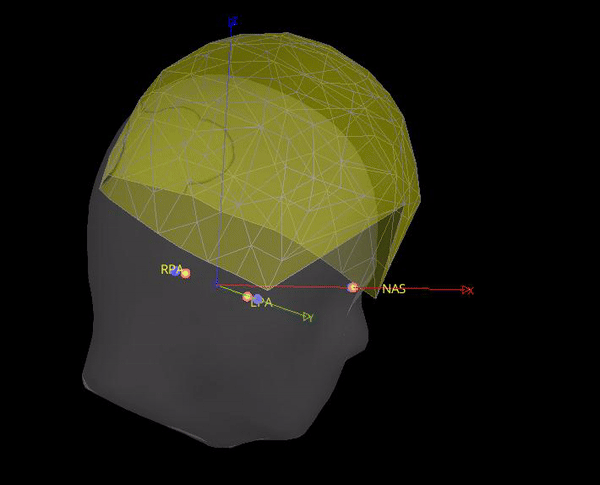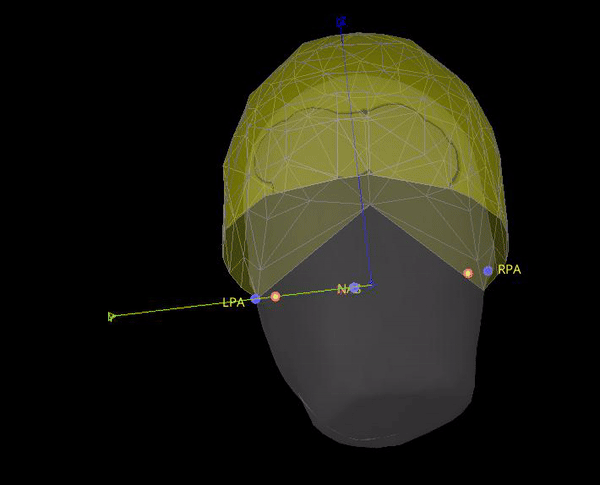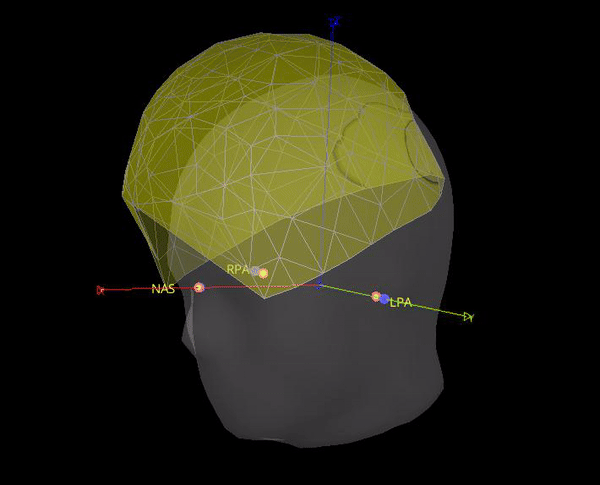
Hi Raymundo,
thank you so much for looking into that!
@Raymundo.Cassani
In theory it makes sense to use the position of the position coils as an approximation to the fiducials. However, the locations that are saved in the .mrk file do not help for the coregistration, they are quite random, check the gif below. Axis are made with the anatomical fiducials. Yellow points (with red border) are the positions of the NAS, LPA and RPA coils according to Yokogawa_file.mrk .
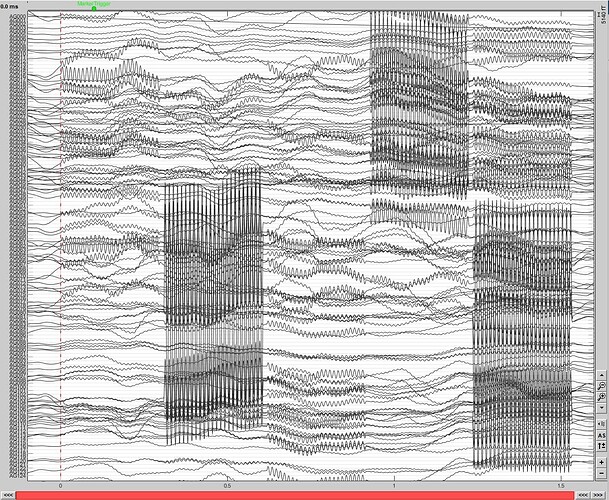
I actually excluded the file I sent you back in January from the analysis in the meanwhile, due to the bad data quality that will also have influenced the coregistration. Sorry about that! This participant had ferromagnetic implants that introduced low-frequency artifacts as you can see here and that probably messed up the positions for that file (-mrk file loaded in brainstorm):
Again, sorry about that - I sent you another file with better data quality. It would be great, if you could rerun this with the new file once to check, if possible?
The txt files do not exist for every single file, so I guess I need to rely on the .mrk files where possible. Do you think it still gives a reasonable approximation of the head localisation? Again- I know that the co-registration is far from optimal in any case if you don't have the exact head points. Another caveat with that data is, that the fiducials were placed a bit differently - i.e. higher above the nasion and more frontal/anterior for RPA and LPA.
Thank you so much for your support,
Bahne
Will you use default anatomy for all the Subjects?
Sounds good, although a case by case approach may be needed.
Do you happen to know the position of the head localization coils on the Subject anatomy?
If so, a better positioning could be obtained by changing the Fiducials in the anatomy for the position of the coils.
With the new shared example files:
Left: Ignoring the information in the .mrk file
Right: Using the information in the .mrk file
Hi Raymundo,
I so far used the subject anatomy and put the positions manually based on where they were placed at that center. I assume the right sided example would be re-assuring, that using the .mrk file to co-register files even without known positions works with my script in theory. I will have to live with overall inaccuracy anyhow, I guess.