Hi @Francois
I did some test, it's working great with Cat12 !!
Just a few comment :
-
The order of the tissue is reversed from what is used in nirstorm. Current version output is 5:scalp, 4:skull, 3:CSF, 2: gray, 1:white matter when NIRSTORM use 1:scalp, 2:skull, 3:CSF, 4: gray, 5:white matter. Is it possible to change that ?
-
It might be that my computer is too old, but even if the segmentation is really fast (<60min), then the importation is also taking ~1hour. It seems that the nearest neighbour search step is taking a lot of time.
-
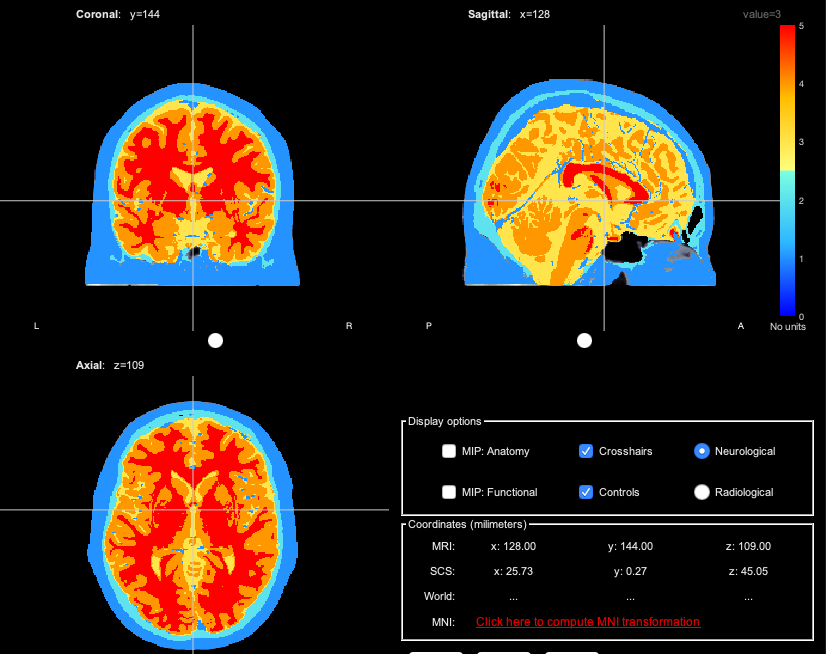
When using the overlay option to display segmentation over MRI, it's actually hard to see the anatomical MRI under the segmentation. And the transparency cursor doesn't change anything.
also, for my personal knowledge, do you think it could be possible to refine this segmentation using an additional T2 image ?
edit : [quote="Francois, post:10, topic:19129"]
Last question: I could get all the p1..6.nii files with your last indications.
With the logic of the code you sent me, I get very noisy separation for the skin (4) and skull layers (5). There are plenty of isolated "skull voxels" in the skin layer (example below: select voxel is classified as skull).
Is this expected, or am I doing something wrong?
Any quick and easy solution for improving this?
[/quote]
Yes, I was worried about that too. but it seems we have the same behavior in the colin27 template that was made by Thomas :
Best regards,
Edouard