Dear Brainstormers,
I am doing a volume source estimation using min-norm and want to define an ROI based on a few MNI coordinates. I generate BEM surfaces, compute head model in MRI volume as the source space, and compute sources using free dipole orientations, using these codes (if helpful):
% Process: Generate BEM surfaces
bst_process('CallProcess', 'process_generate_bem', , , ...
'subjectname', SubjectNames(i), ...
'nscalp', 1922, ...
'nouter', 1922, ...
'ninner', 1922, ...
'thickness', 4);
% Process: Compute head model
bst_process('CallProcess', 'process_headmodel', sFilesAvg(1), , ...
'sourcespace', 2, ... % MRI volume
'eeg', 3, ... % OpenMEEG BEM
'openmeeg', struct(...
'BemSelect', [0, 0, 1], ...
'BemCond', [1, 0.0125, 1], ...
'BemNames', {{'Scalp', 'Skull', 'Brain'}}, ...
'BemFiles', {{}}, ...
'isAdjoint', 0, ...
'isAdaptative', 1, ...
'isSplit', 0, ...
'SplitLength', 4000));
% Process: Compute sources [2016]
sAvgSrc = bst_process('CallProcess', 'process_inverse_2016', sFilesAvg, , ...
'output', 1, ... % Kernel only: shared
'inverse', struct(...
'Comment', 'sLORETA: EEG', ...
'InverseMethod', 'minnorm', ...
'InverseMeasure', 'sloreta', ...
'SourceOrient', {{'free'}}, ...
'Loose', 0.2, ...
'UseDepth', 0, ...
'WeightExp', 0.5, ...
'WeightLimit', 10, ...
'NoiseMethod', 'median', ...
'NoiseReg', 0.1, ...
'SnrMethod', 'fixed', ...
'SnrRms', 1e-06, ...
'SnrFixed', 3, ...
'ComputeKernel', 0, ...
'DataTypes', {{'EEG'}}));
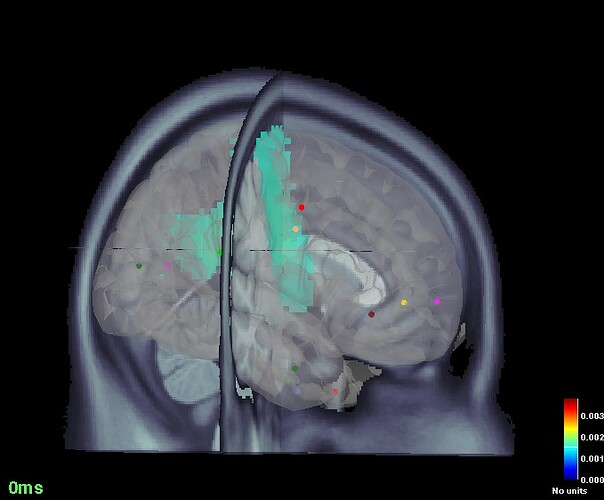
Once I get the sources,I open the figure showing cortical activations on 3D MRI and using Scouts panel in BS GUI I define new volume scouts using MNI coordinates.
The size of the sources matrix I see in imaging-kernel turns out to be 13478. Now my problem is that for certain MNI coordinates, it falls in error saying it cannot create such scout because it goes beyond the size of the vertices. I thought maybe it falls out of the brain, but I see it doesn't. Like, it can define [-52 11 41] which is close to the brain border, but cannot generate [-3 27 -9], which lies somewhere at the center of the brain.
Bellow is a picture of the error I get in interface:
I appreciate any help in advance.
Maryam