Hello, when I computed the source [2018], I faced the following problem:
--------------------------content in the command window-------------------
BST_INVERSE > Depth weighting is only available for minnorm, ignoring option UseDepth=1
BST_INVERSE > Rank of the 'EEG' channels, keeping 59 noise eigenvalues out of 60 original set
BST_INVERSE > Using the 'median' method of covariance regularization.
BST_INVERSE > Covariance regularized by flattening tail of eigenvalues spectrum to the median value of 1.0e-05
BST_INVERSE > Using 'free' surface orientations
Error using svd
Input to SVD must not contain NaN or Inf.
Error in bst_inverse_linear_2018 (line 729)
[Ua,Sa,Va] =
svd(L{kk}(:,ndx),'econ');
Error in process_inverse_2018>Compute (line
645)
[Results, OPTIONS] =
bst_inverse_linear_2018(HeadModel,
OPTIONS);
Error in process_inverse_2018 (line 24)
eval(macro_method);
Error in panel_protocols>TreeInverse (line
1224)
[OutputFiles, errMessage] =
process_inverse_2018('Compute',
iStudies, iDatas);
Error in panel_protocols (line 44)
eval(macro_method);
Error in
tree_callbacks>@(h,ev)panel_protocols('TreeInverse',bstNodes,'2018')
(line 2436)
gui_component('MenuItem', jPopup, ,
'Compute sources [2018]',
IconLoader.ICON_RESULTS, ,
@(h,ev)panel_protocols('TreeInverse',
bstNodes, '2018'));
--------------------------content in the command window-------------------
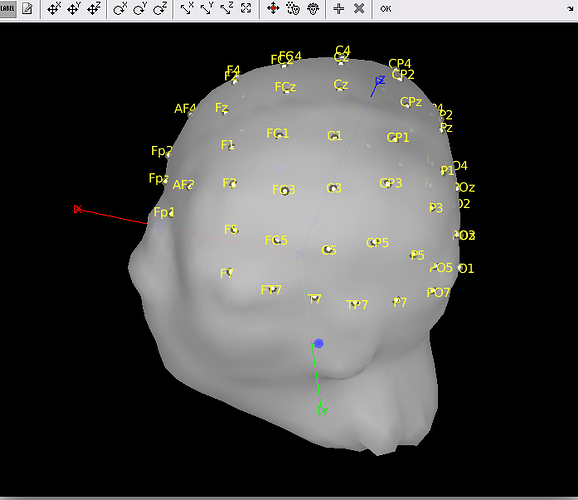
PS: I used the individual MRI and the view seemed ok.
I used the neuroscan quickcap 64 channel electrode template (M1 M2 CB1 CB2 were deleted resulting 60 channels remained.) and used the openMEEG BEM model.
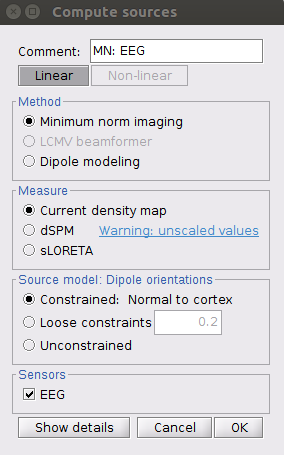
For source computing, since I was analyzing the resting EEG data, I just used the Identity matrix as the noise covariance matrix and made all other settings default.
(Under this condition, I think the source computing is not related to the EEG signal if I didn't calculate something like the data covariance matrix. Am i right? I am not very sure.)
At the same time I also checked the following issues to see whether it was caused by the existence of zero or nan in headmodel.Gain:
-------check part-----------
Data exported as "HeadmodelMat"
size(HeadmodelMat.Gain)
ans =
64 45009
numel(find(HeadmodelMat.Gain==0))
ans =
0
numel(find(HeadmodelMat.Gain==nan))
ans =
0
----------------check part---------------------
So may I know what I should do next and anything I should check?
Looking forward to your kind reply and thanks in advance.