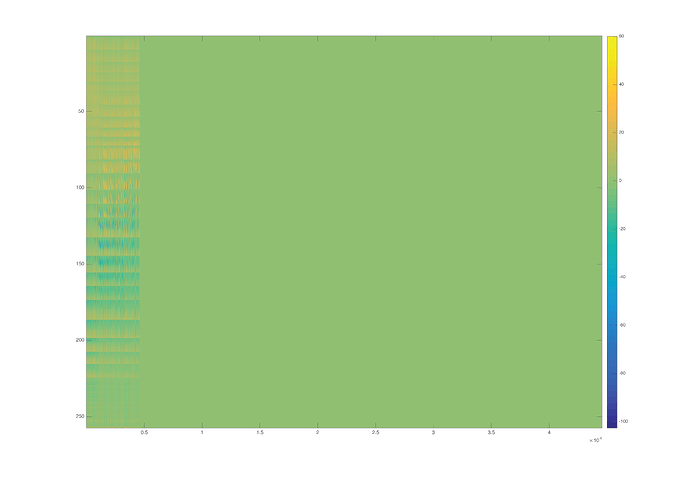
Yes I used OpenMEEG, and yes there are many zeros in the gain matrix, thought that was suspicious:
Data exported as "HeadmodelMat"
>> size(HeadmodelMat.Gain)
ans =
257 44565
>> numel(find(HeadmodelMat.Gain==0))
ans =
10261496
>> figure;imagesc(HeadmodelMat.Gain);colorbar
I will report the issue to the OpenMEEG team, and try the 3-shell sphere model.
Karl