To better illustrate what I'm trying to explain here, please see this simple example protocol, that you can load with the menu File > Load protocol > Load from zip file.
TutorialEpilepsy_test_flip.zip (707.0 KB)
- 01: I took one second from the epilepsy tutorial
- 02: Right-click on the channel file > Edit channel file, and I edited the channel labels to flip left-right (e.g. C3<=>C4, Cz unchanged)
- 03: I edited manually the positions of the channel file in 02, with the following script, so that in appearance, the channel files in 01 and 03 look the same when displayed on the scalp, but with a different channel order:
for i=1:length(c01.Channel)
iChanLoc = find(strcmpi(c02.Channel(i).Name, {c01.Channel.Name}));
c03.Channel(i).Loc = c01.Channel(iChanLoc).Loc;
end
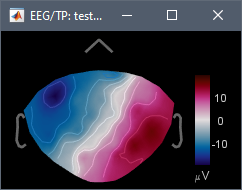
- 04: Average of data in 01 and 03: channel file shows all the electrodes on the midline (indicating that the positions where correctly flipped), but the EEG data is unchanged (the F matrix was not flipped)
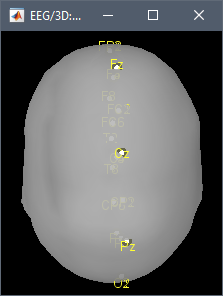
- 05: Result of selecting data 01 and data 03 in Process1 and running "Uniform list of channels > Use the first channel file in the list" => This uses the original channel file, but the topography is flipped left-right with respect with 01.
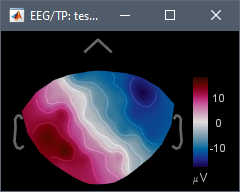
- 06: Average of data in 01 and 05: channel file is identical in both and therefore unchanged, data file is average with its flipped version, resulting in a symmetrical left-right topography.
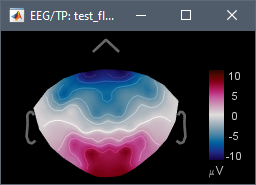
What you should get for each subject is what is 05, before averaging across subjects.