after that connectivity ==> Coherence AxB
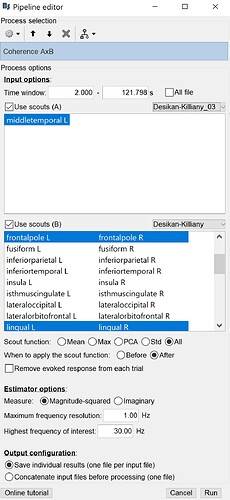
If you select the option "Scout function: All", it will keep the values for all the possible pairs of sources within all the selected regions. This is probably not what you are trying to do if you want to consider these as "regions of interest".
when I want to do T.Test between this two Coh for analyzing the connectivity between my regions during rest state and task state I can see just Difference of means. What' is the problem?
A t-test is comparison of two groups of samples. Here you have only one sample for each measure and each condition (one experimental session, one subject). The only thing you can do is to compute a different of your two samples, you will not be able to get any significance measure associated with the result of this difference.
Before trying to do this on your own data, please read the corresponding tutorials:
http://neuroimage.usc.edu/brainstorm/Tutorials/Difference
http://neuroimage.usc.edu/brainstorm/Tutorials/Statistics
I can't display Coh as a graph because of an error
Probably because of the option "Scout function: All" you selected. This is not supported by most of the display functions. Note that this graph will not help you understanding anything if your connectivity matrix is not thresholded by some form of statistical testing. For your raw connectivity matrices, I recommend you keep on looking only at the full matrices, it will prevent you from false interpretations of the graphs.
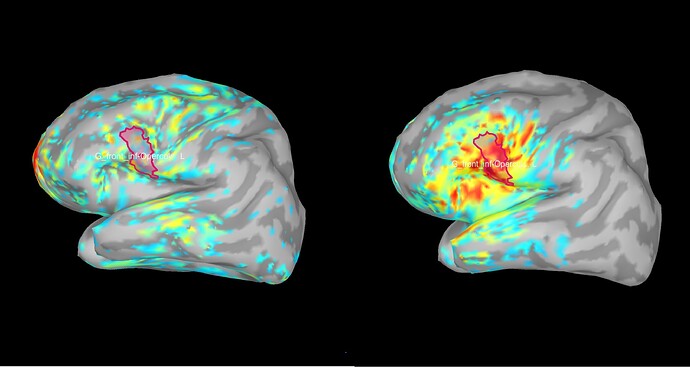
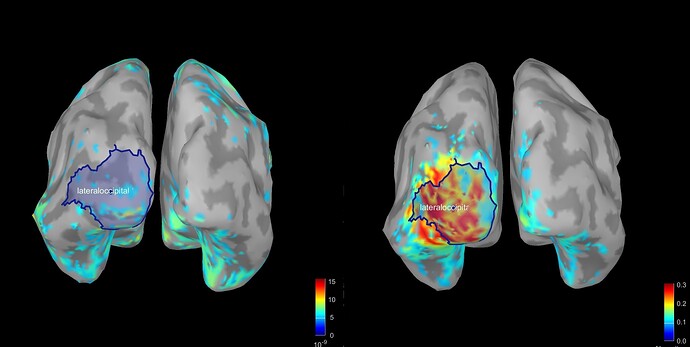
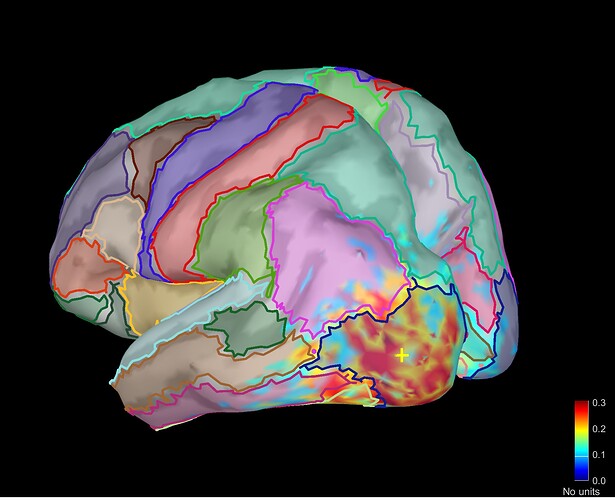
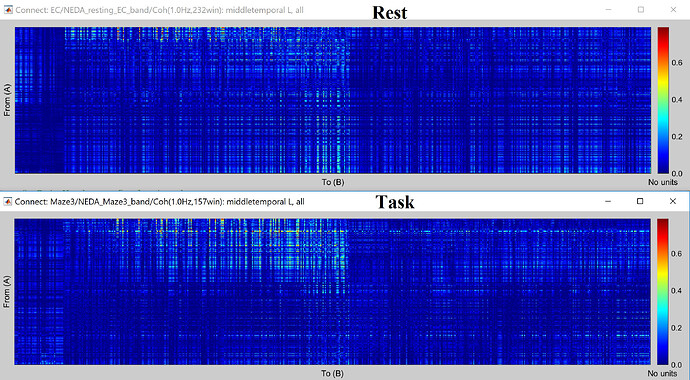
when I display Coh as an image I have this following figure. How can I interpret this figure exactly?
This is the connectivity measure you selected (magnitude-squared coherence) between all the pairs of sources within the scouts you selected, for the frequency that is currently selected in the Brainstorm window (frequency slider).
How can I use these two Coh matrices and how can I say which rest or task state has more connectivity between my ROIs.
Compute a difference between the two.
To know which of the values in this difference are significant, you need many subjects (or many repetitions of the same experiment on the same subject).