Hi Raymundo,
I came across another instance that I wanted to ask you about:
For some of our participants, we could not record CapTrak data due to mechanical issues. In these instances, we use the channel file from the .set file, which has the updated BrainVision standard 96-channel cap layout.
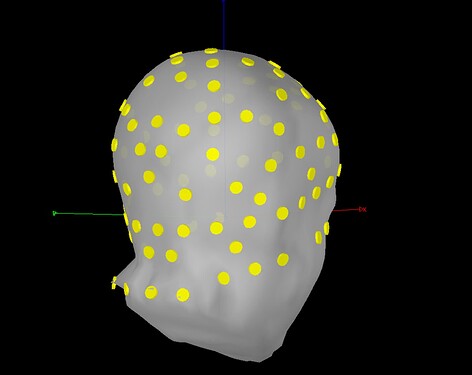
On first import, I refined the channel locations to the digitized head points, resulting in the following registration:
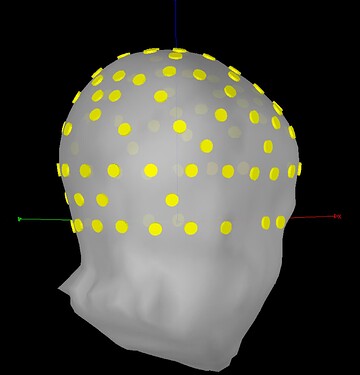
In trying to troubleshoot this issue, I re-imported the data without refining the channel locations to the digitized head points of this person's MRI, leading to this registration (which I think is better than the first one):
If I refined the channel locations to digitized head points for other subjects that don't have CapTrak data, but did not do so here, is this an issue? I'm learning Brainstorm and want to ensure changing this setting in this way doesn't affect reproducibility in methods between subjects.
Thank you,
Adam