Please send me your .pom file.
Attached.
all.pom (14.0 KB)
If I right-click on an empty folder > Import channel file, select file format "EEG: Curry" and then change the type of the channels to SEEG, I get this:
So I guess there is nothing wrong with this .pom fil or the Brainstorm .pom reader.
How are you trying to import this file?
Thanks so much, now I can import. I guess I didn't change the format to EEG: Curry before (but I could see and select the file!)
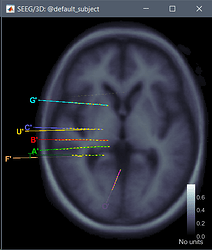
Anyway, there is another issue. Here is what I get.
The problem is that based on the MNI locations in .pom file and other resources the electrodes are implanted in the left hemisphere. Also, the O' electrode should be in frontal region (Y and Z are positive).
can you realize the source of the problem or I'm missing something?
Thanks
There is obviously something wrong with the transformations applied to register the SEEG coordinates and the anatomy. This can have 3 origins:
- You`re not using the appropriate MRI or not loading it correctly: your MRI volume does not include the world transformation needed to interpret correctly the positions in your .pom file or you answered NO when asked whether you wanted to use it (https://neuroimage.usc.edu/brainstorm/CoordinateSystems#World_coordinates)
- The coordinates in the .pom file were not generated to fit this MRI volume but another one
- There is a missing standard transformation in the .pom file reader (https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/io/in_channel_curry_pom.m)
I never heard back from @sathompson8 so I could not investigate any possible bug in the reader. At that time, I didn't have the specification for this file format, I don't know in what referential the files are supposed to be saved.
Maybe you can investigate this a bit further if you have full datasets in hand.
Thanks for your complete response.
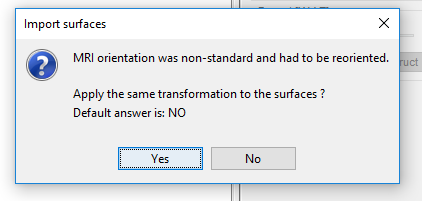
So, apparently MRI and Surface are not registered and aligned correctly in my data-set and it seems they use non-standard orientation (No matter how I respond to this question it won't be fixed).
I tried four different approaches for Anatomy.
- Manually aligned surface and MRI (while their axes are not?!)
- Generated a surface from the MRI and ignore the current surface,
- Just Imported the surface and Ignored the MRI
- Used the default anatomy.
Nevertheless, in none of them, I could see the correct electrode positioning. The MRI has vox2ras matrix.
The electrode coordinate in the .pom file looks like MNI coordinate. however, as I said, I cannot see those locations in MRI views, even for the default anatomy. Can it be related to in_channel_curry_pom.m function?
This is the important part of the code:
ChannelMat.Channel(iChannel).Loc = [-xyz(2); xyz(1); xyz(3)];
Thanks
There is probably a transformation missing in the function in_channel_curry_pom.m.
You can check out other in_channel_* functions for examples.
There is also something wrong in the import of your surface, obviously.
All the import functions (in_tess*) have transformations added to them, but that are valid only in one specific context (FreeSurfer or BrainSuite output folder typically). Importing randomly a surface in a random format has very little chance to end up where you want it.
If these surfaces were generated with Curry, then you may need to add an entry "Curry surface" with the appropriate transformation, and make sure it matches the standard procedure that any Curry user would use.
Coordinate systems and transformations are a never-ending nightmare...
I can help you with this, but not immediately, I'm leaving tomorrow for a conference and won't be back before the end of next week.
Hello everyone,
Having read the posts above, I was wondering if there is a general pipeline to import .pom files from Curry into BST?
Thanks!