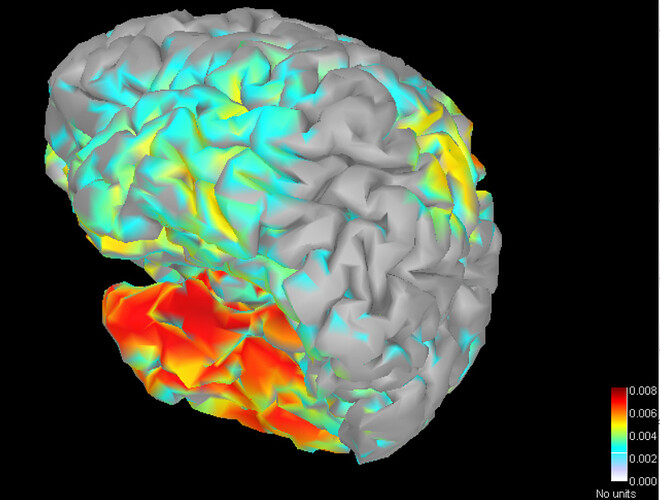
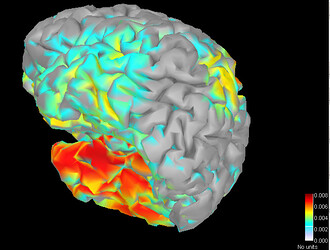
Is there any possibility I can get the spatial coordinate of the dipoles after the source imaging?
If you used a dipole fitting or dipole scanning procedure, you just have to click on a dipole to get its coordinates:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutDipScan#Displaying_dipoles
If you are using distributed source models, you can get coordinates of any point on the cortex surface with the popup menu "Get coordinates":
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy?highlight=(Get+coordinates)#Get_coordinates
Thank you very much, I am very honored to be replyed.
I have already got the spatial coordinate of the dipoles so far, and I am wondering how can I get the 3-D pictures with the 15,002 dipoles' spatial coordinate? Is there any program that I can achieve that?
Hello,
I have got the 15,002 dipoles' spatial coordinate(15002x3 matrix).But I want to transform it into a three-dimensional numerical matrix with spatial physical information.Is there any chance I can get that?
Thanks for your time.
Wish you a good day.
The 3D coordinates of the vertices of the cortex surface are available in the corresponding .mat file, in the field Vertices:
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_Surface
I'm not sure I understand what kind of display you expect.
Could you post an example image?
When I was performing this step:
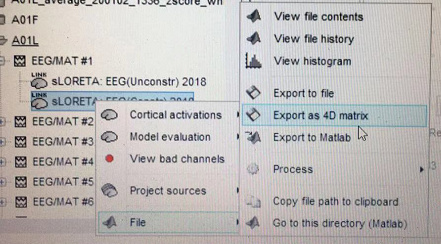
I got a 3D matrix named Cube(72x92x62) such as this:
![]()
And my problem is how do you get the 3D picture like this:
Is there any relationship with the Cube I export?
If is not, how do you get the 3D picture of all the dipoles? Or what is the program of getting the 3D picture?
This is really a big question to me, I am really looking forward to your reply.
Thank you very much.
This menu "Export as 4D matrix" interpolates the 15000 values obtained on the surface in a volume (similar to what you obtain with the menu "Cortical activations > Display on MRI (MRI viewer)", and saves this volume to a .nii file. This is unrelated with the figure you included in this message.
Do you mean that you want to find a way to create this 3D figure from a script instead of using the GUI?
You can probably do this with the function view_surface_data.m or the script script_view_sources (read the header of the files for additional help):
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Reference:_Display_functions
You can find examples in the script reproducing the introduction tutorials:
https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_introduction.m#L640
Or you can use the process "File > Snapshot"
When I was performing this step“Cortical activations> Display on cortex”, and continue to “Get coordinates”
I was getting some spatial coordinate of the dipoles, and each dipole has a value.
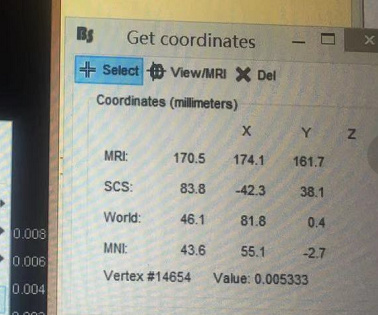
I am curious how do you save the [x,y,z,Value] ( Do you save it as a martix or what?)and where do you save it?
The locations of the surface vertices are saved in the surface files (SCS coordinates):
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_Surface
The other coordinates are computed from these SCS coordinates with the function cs_convert:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems#Converting_between_coordinate_systems
Sorry for my poor English, I might not make myself clear.
I would like to find the value of each dipole after computing the sources.
In this way, each cooridnate has a value.
For example, in that picture, when x=83.3 y=-42.3 z=38.1 the value is 0.005333.
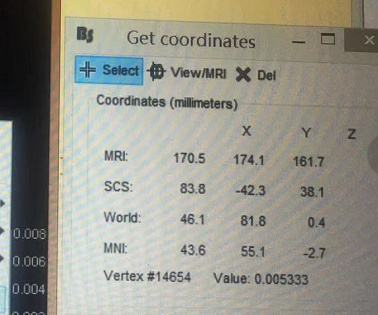
First, where should I find the value?
Second, does the Cube( after the step"Export as 4D matrix" ) exist for this 3D picture?
Thank you for both your time and your patience again.
The locations are in the cortex surface (field Vertices):
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_Surface
The values at surface level are in the source file (field ImageGridAmp, if you selected the advanced option "Full sources"):
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive
The values re-interpolated in the cube must be recomputed explicitly (eg. with the menu "Export as 4D matrix")