Thanks for that answer Raymundo,
What I want to achieve has been answered here.
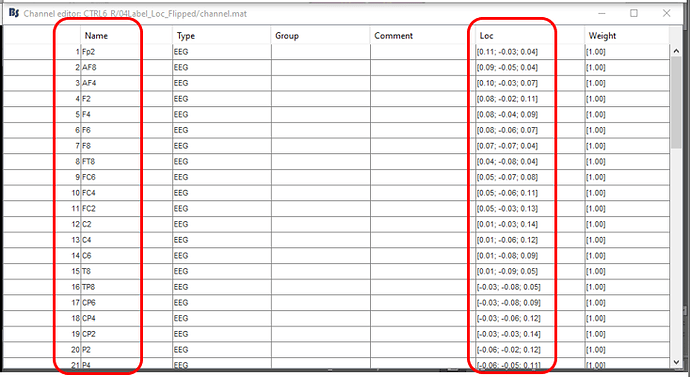
Basically, I would like to flip the EEG layout from right to left (both in channel label order and the locations). The effect that I would like to get eventually is the first image shown above.
What I hoped is by using the attached custom made BioSemi 64 channel position file, that I would be able to get the EEG channel layout shown by the photo. But when you import the attached channel position file, it is not showing the desired effect shown below (even though the channel position file was made correctly to represent the below photo).
So I am looking for ways to import a channel position file that will immediately reflect what I am showing down below as a photo.
Hope that made sense.