Hi François,
Thanks for the information. I'm not sure why, but at first the GUI was not giving me the option to project scouts from one subject kernel to another - or any other kind of projection for that matter - via the scout tab. So, instead, I decided to drop my 5 subjects into Box 1 and project the sources onto the default anatomy via Run > Sources > Project to default anatomy. This took a long time and produced 5 very large group analysis files. But, for some reason, after doing this I now do see the option to project across kernels as shown in your snapshots, regardless of whether I open one of the original surface kernels or one of the massive group-analysis surface files. Can you explain this or point me to a tutorial that would help me understand?
In any case, the good news is that after this procedure I was then able to project the Desikan-HMAT hybrid atlas from one of the original subject kernels to another using the Scout tab path shown in your snapshots. The projected ROIs look good now, but I don’t want to have to project every new subject (or any subject, actually) to the default anatomy template just to get the Scout tab option you say I should have by default. Moreover, you advised me against projecting Desikan scouts across kernels because these scouts are already in subject space, which makes a lot of sense. So, I’m thinking that the best approach would be to automate the scout selection and hybrid atlas generation for each participant via a script that I could add to my pipeline.
Put more formally: I want to generate a script which automatically creates a user atlas containing the same very specific scouts from two standardized atlases, so that I can iterate it through my sample. I could of course do this manually for every participant brain kernel, but I’m dealing with a resting state protocol here with around 200 participants, so I strongly prefer to automate this. I already know how to generate and concatenate .m scripts derived from processes done via Box 1, but this scout selection and atlas generation procedure is all done via the Scout tab, so I’m not sure how to go about generating this script I need.
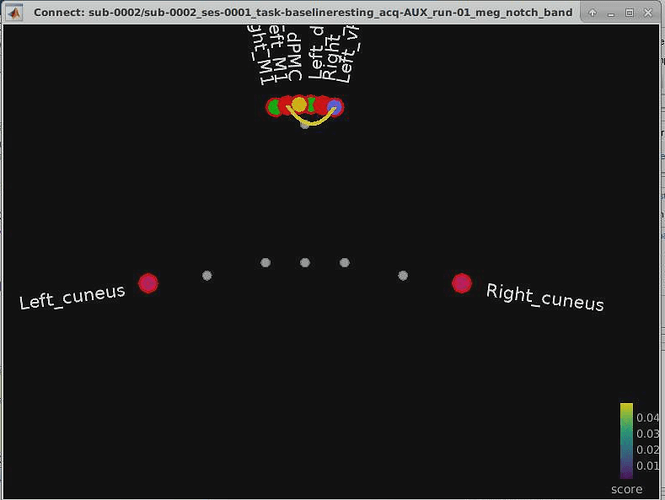
While we’re on the topic, I also wanted to ask you a smaller question about scout labels and tags, because they’re giving me some trouble when I look at the results of my NxN imaginary coherence connectivity matrix in graph form. As you can see in the ugly image attached, the HMAT scouts are all piled together in the center and the two Desikan scouts are panned to either side based on their “hemisphere tags”. By this I mean the two-letter acronym (for instance: LO = Left Occipital) which can be seen to the left of the scout names in the atlas list under the scout tab itself. My question is: how can I add these “hemisphere tags” to the HMAT atlas scouts, or even remove them from the Desikan atlas scouts in order to resolve this visualization problem? Renaming the scouts via Scout tab > Rename doesn’t work, so I’m out of ideas.
Many thanks in advance,
Oscar