Hello
i’m trying to work with eeg simulation, already i could simulate my eeg from some specific scouts with the process menu:
sFiles = bst_process(‘CallProcess’, ‘process_simulate_matrix’, …
sFiles, [], …
‘subjectname’, SubjectNames{1}, …
‘condition’, num2str(i), …
‘samples’, 150, …
‘srate’, 100, …
‘matlab’, ‘Data(1, = 0.0008sin(4pi*(t-5)).exp(-(4t-10))’);%%
= 0.0008sin(4pi*(t-5)).exp(-(4t-10))’);%%
%Process: Simulate recordings from scouts
sFiles = bst_process(‘CallProcess’, ‘process_simulate_recordings’, …
sFiles, [], …
‘scouts’, {eval(‘cortexFull.Atlas(1,8).Name’), {num2str(i)}}, …
‘savesources’, 1);
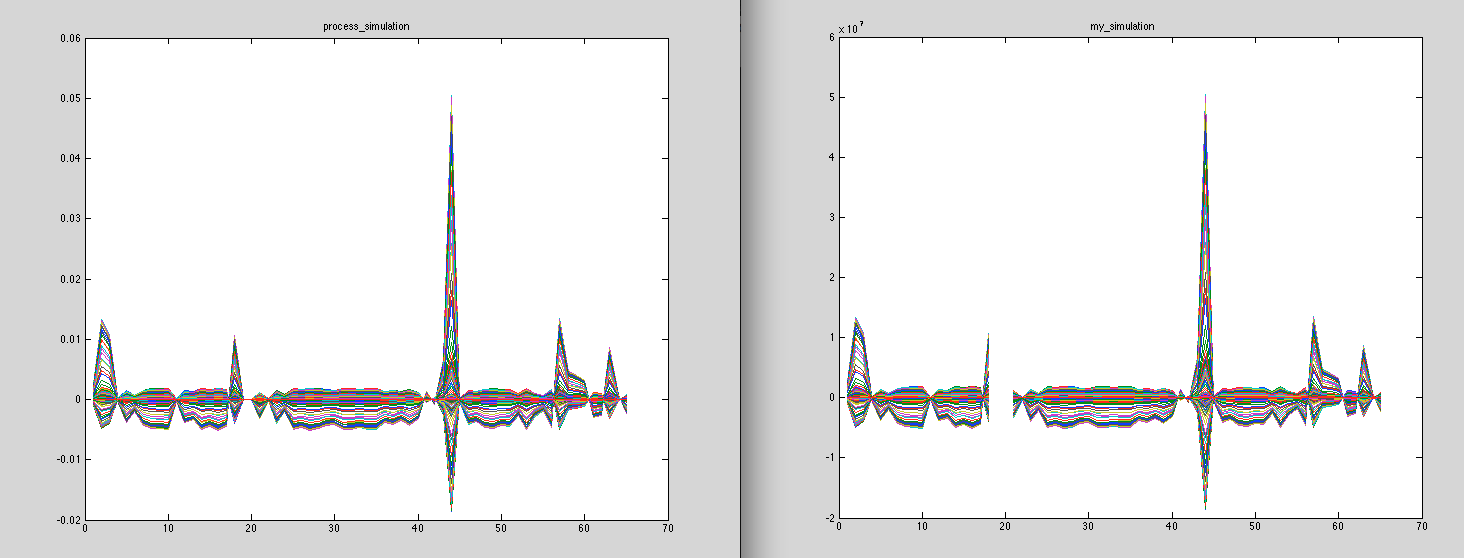
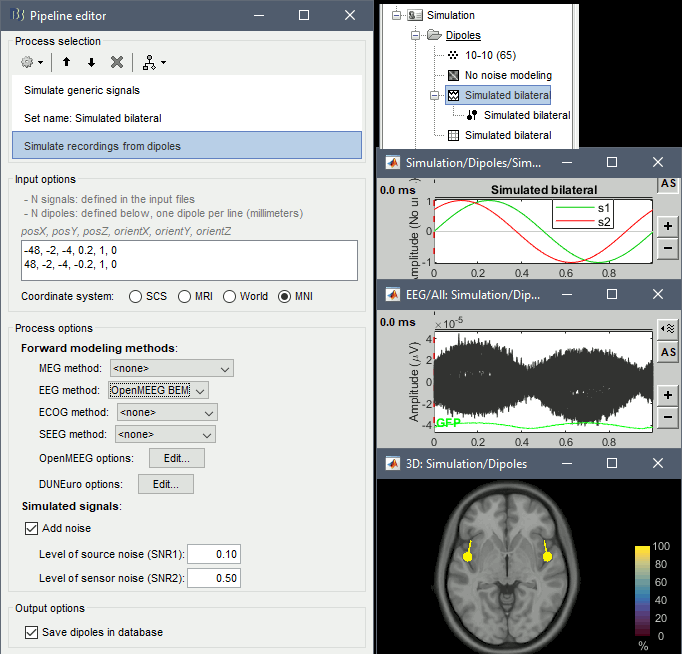
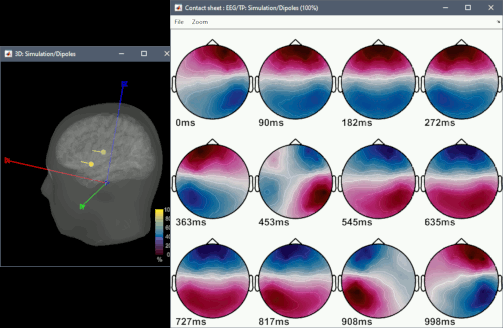
But i would like to know more detail about how the software use the information of the head model (my lead field), to generate the eeg recording for the specifics electrodes.
I would like to know every detail that permit me understand the basics behind this magic process, where i generate the specific source position (vertex) and some generic signal, and magically appear a eeg scalp record.
Saludos desde Chile