Hello,
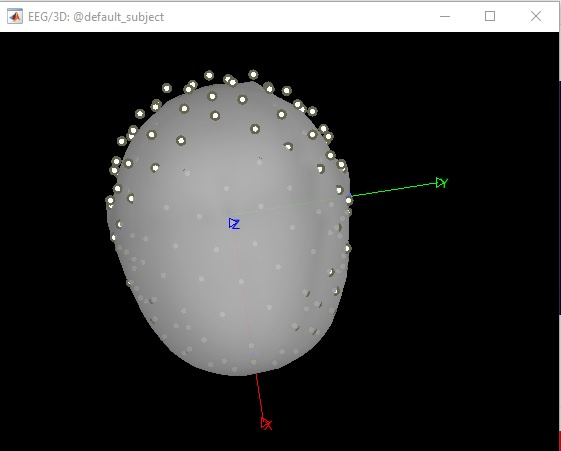
I have EEG data collected by using EGI's 128-channel system. When I imported the data in mff format, I noticed that channel locations were not accurate as you can see below.
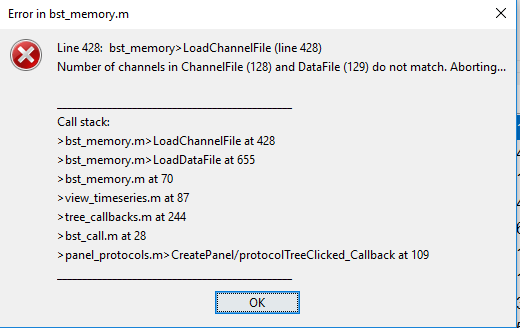
So I tried different EEG default cap options and ran into the following problems
-
GSN 128:
-
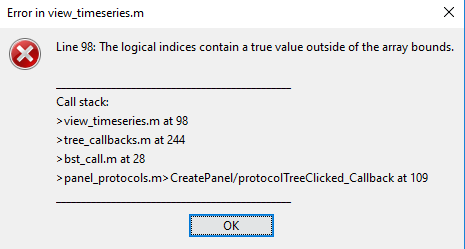
- GSN Hydrocel 128 E001 and E1:
Can you tell me how to troubleshoot?
Sorry for the reponse delay.
I have EEG data collected by using EGI’s 128-channel system. When I imported the data in mff format, I noticed that channel locations were not accurate as you can see below.
Is this a template anatomy?
Are you expecting to get 3D positions from the .mff file, or are you planning to use default EEG positions from Brainstorm?
If you are using only Brainstorm templates, you should obtain electrodes positioned exactly on the head surface. In all the other configurations, it is normal not to have the two surfaces matching. In this case, you need to edit this registration manually:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Register_electrodes_with_MRI
So I tried different EEG default cap options and ran into the following problems
If you want to add default electrode positions to you dataset, you should first link your recordings to the database, then right-click on the channel file > Add EEG positions.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Prepare_the_channel_file
Here you apparently deleted your channel file and replaced it with one that did not match.


