Dear Francois,
I tried to run the Epileptogenicity process on one of my seizures, following the instructions in the tutorial, however using different latencies: I first imported a 15-sec baseline, and a 50-sec seizure epoch, consisting of -10 sec prior to onset (preictal) and 40 sec after (ictal). I used various latency strings using some frames with negative time window values ('-10:2:36','-10', '-10:2:-2'); when I did this, I either got this error,
** Error: [process_epileptogenicity] Epilepsy > Epileptogenicity maps (A=Baseline,B=Seizure)
** Line 212: Index exceeds array bounds.
**
** Call stack:
** >ImaGIN_Epileptogenicity.m at 212
** >process_epileptogenicity.m>Run at 277
** >process_epileptogenicity.m at 28
** >bst_process.m>Run at 231
** >bst_process.m at 36
** >panel_process2.m>RunProcess at 150
** >panel_process2.m at 26
** >gui_brainstorm.m>CreateWindow/ProcessRun_Callback at 772
** >bst_call.m at 28
** >gui_brainstorm.m>@(h,ev)bst_call(@ProcessRun_Callback) at 296
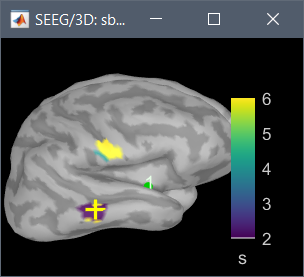
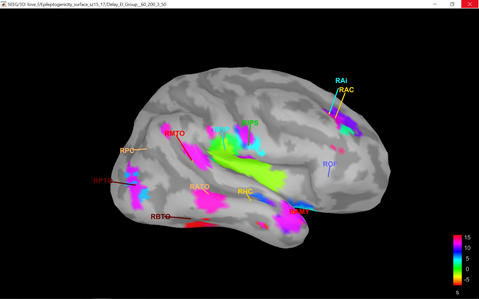
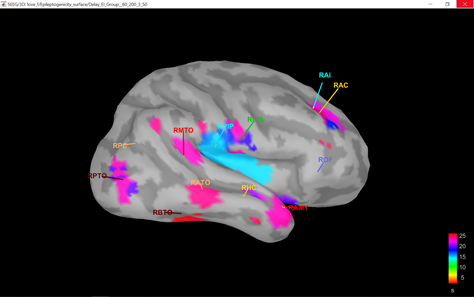
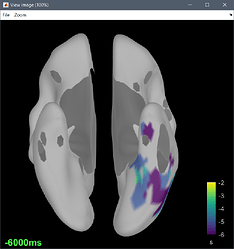
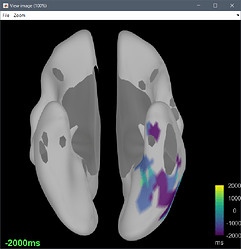
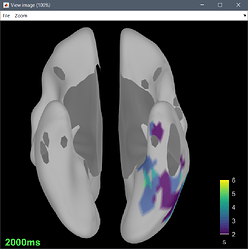
or I got through, but the delay map pictures start at frame 0, instead of -10. It seems that the code is built to analyze seizures from frame time 0 and on.
It would be desirable to adjust the code to be able to accommodate negative frame times (prior to seizure onset (0sec) time; otherwise, one would have to add markers at (mrk = eo - 10 sec) in the raw link file, then import in database using mrk+ 50 sec, which is doable but takes some time if one has 15 seizures to process per subject (with my recordings, I can assume this is not singular, the seizure onset marker eo was added in the original file, prior to import into Bst). It would be more natural to keep seizure onset as time/frame 0 sec, import a -10:40 sec epoch into database, then run the code maintaining this time window designation. The reason one does not want to start with t=0 sec at times is that there is some uncertainty or interrater divergence (in some cases) as to where the clinical seizure onset is, so one may want to see the ictal discharge in evolution, with some buffer pre/post onset.
I think the main change would be starting a line 198 in imaGIN_Epileptogenicity.m, as follows:
> for i0 = 1:size(DD,1)
> if (length(Horizon) == 1)
> TimeWindow = min(latency(:)) : TimeResolution : Horizon+1+max(latency(:));
> elseif (length(Horizon) > 1)
> TimeWindow = min(latency(:)) : TimeResolution : Horizon(i0)+1+max(latency(:));
> end
with min(latency(:)) replacing 0.
There may also need to be some other minor downstream changes, since the picture of the delay map still shows a legend 0:36 sec, as opposed with -10:36, after the modification above.
I will attach a subject's folder via email with one baseline and seizure epoch as described above, please try it with latencies as above ('0:2:36','-10:2:36','-10', '-10:2:-2') to see what I am talking about.
Thank you much, as always,
Octavian