What's new
May 2011
Brainstorm reference paper
Publication of a special issue of the journal Computational Intelligence and Neuroscience: "Academic Software Applications for Electromagnetic Brain Mapping Using MEG and EEG". The article describing Brainstorm will now be used as the its reference paper, please cite it in your publications (see How to cite Brainstorm).
Tadel F, Baillet S, Mosher JC, Pantazis D, Leahy RM, “Brainstorm: A User-Friendly Application for MEG/EEG Analysis,” Computational Intelligence and Neuroscience, vol. 2011, Article ID 879716, 13 pages, 2011. doi:10.1155/2011/879716 [ html, pdf ]
Database explorer
The file manager has been extended to support drag'n'drop and copy-paste operations. You can now move or copy easily the files from a condition or subject to another with the mouse, using the keyboard shortcuts CTRL+C (copy), CTRL+X (cut) and CTRL+V (paste), or the File section in the popup menu.See tutorial: ?First steps.
Warping default anatomy
It is now possible to create pseudo-individual anatomies for the subjects for which you do not have any anatomical MRI. This operation is possible only if you have acquired some head points using a tracking system (eg. Polhemus Isotrak). The default anatomy (MNI/Colin27) is transformed to match those match those head points. See tutorial: Warping default anatomy.
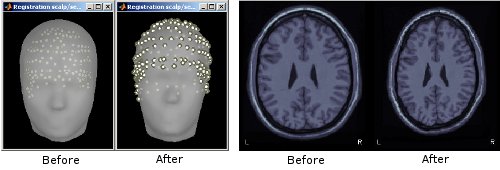
Refine registration using head points
When importing MEG recordings, the registration between the sensors and the anatomy was based until now only on three points (nasion, left ear, right ear). This registration can be very imprecise in some cases. If you have digitized many head points with a tracking system before the recordings acquisition, the registration can be refined by trying to minimize the distance between the scalp surface and those digitized head points. This operation will be performed automatically for new datasets, but can also be called manually: right-click on channel file > MRI registration > Refine registration using head points. See tutorial: Refine registration using head points.
April 2011
BEM head model for EEG users
Great news for the EEG users, a new option is available for the computation of the head model: OpenMEEG BEM. This forward model uses a symmetric boundary element method and was developed by the French public research institute INRIA (website). It uses three realistic layers (scalp, inner skull, outer skull) that can be generated in Brainstorm. It provides for EEG a much better forward solution than the previous "3-shell sphere" model. EEG users should try to switch to this method quickly. It is not necessary for MEG users, as the "overlapping spheres" method gives very similar results but much faster. See tutorial: BEM head model.

Source estimation for the full brain volume
The head models computed with Brainstorm were until now limited to the cortex surface. This choice was following the assumption that most of the magnetic and electric activity that we record outside of the head comes from the cortex. It was a serious limitiation for users who wanted to study some deeper regions and use their own source spaces. To extend the range of analysis possible with Brainstorm, we have added the possibility to construct dipole grids that sample the full brain volume, and the appropriate visualization tools. See tutorial: Volume source estimation.
March 2011
Combined MEG/EEG source reconstruction
The way the contributions of MEG and EEG information are balanced in the combined source estimation has been largely improved. However, we still consider this feature as very experimental and recommend the average user to reconstruct the sources for EEG and MEG separately.
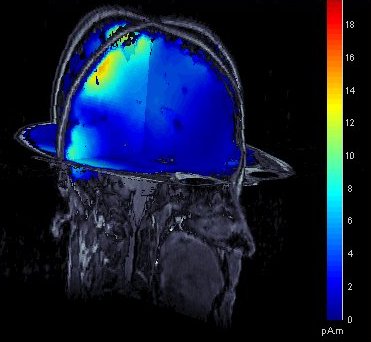
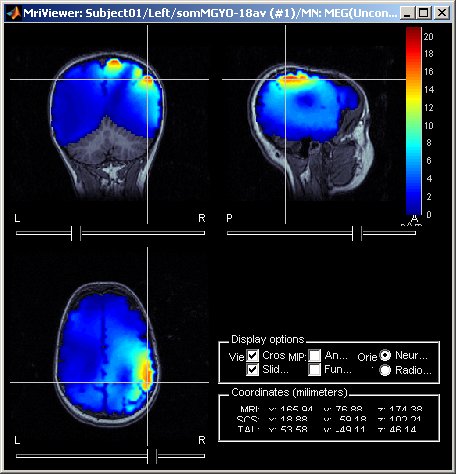
MRI displays: Contact sheets and smoothing
Many new tools for the visualization of sources on MRI slices. First, the sources values are smoothed in space after being re-interpolated in the MRI, which gives nicer and more readable images. The size of the smoothing kernel can be configured: right-click on the figure > MRI display. Then, those images can be used to create contact sheets along all the directions, either in time or in volume. See tutorial: ?Source estimation.
December 2010
Statistical tests: correction for multiple comparisons
When displaying the results of a t-test, there is now a new tab "stat" that is shown in the main window, that allows to changed dynamically the type of correction for multiple comparisons and the threshold.
Graphical batching interface
A completely new version of the "Process" tab is available. It allows the creation of a full processing pipeline in a few clicks, that can be saved or exported as a Matlab script. The processes that are available in the interface are now written in a plug-in way, to make it much easier the future development of new processes. See tutorial: ?Processes.
September 2010
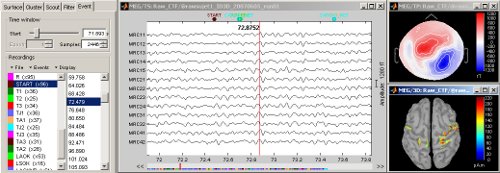
Continuous/RAW file viewer and event editor
Brainstorm can now be used to visualize and process full length continous/raw files in native format. See tutorial: ?Review raw recordings and edit markers.
Detection of bad trials / bad channels
New process available to detect bad trials or bad trials based on peak-to-peak values. The peak-to-peak threshold can be set by channel type.
Estimation of the noise covariance from continuous files
The interface that computes the noise covariance matrix can be called on continuous recordings. Right-click on link to raw file > Noise covariance.
Import/export protocols and subjects
New menus to help users exchange data easily. A full protocol can be exported as a single zip file (menu File > Export protocol), and then imported in another database (menu File > Load protocol > Load from .zip file). Same thing for a single subject: right-click on the subject > File > Export subject. The subject .zip file is actually a full protocol zip file with only the exported subject, hence it should be loaded as a protocol.
June 2010
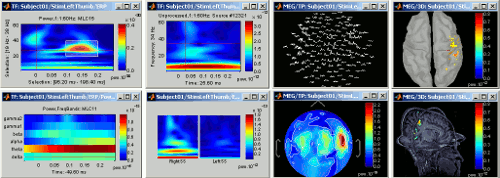
Time-frequency
Computation and visualization of time-frequency decompositions of MEG/EEG and source signals using Morlet wavelets. See tutorial: ?Time-frequency.
Brodman and Tzourio-Mazoyer atlases
Two lists of scouts have been created for the default anatomy (MNI/Colin27), inspired from the atlases of Tzourio-Mazoyer and Brodman. To access those files: display the cortex surface of the default anatomy, then in the "Scout" tab click on the "Load scouts" button, on the right of the scouts list.
Recordings visualization: 2D Layout
The 2D Layout display for recordings has been re-writen to include many tools: sensors selection, interactive gain change, zoom in time and space... See tutorial: ?Exploring the recordings.
March 2010
Support for Xfit dipoles files
Dipoles localizations from the Neuromag Xfit software can be imported in Brainstorm database and displayed on the same figures as Brainstorm cortical maps. See tutorial: ?Import and visualize dipoles from Neuromag Xfit.
Minimum norm solution
Integration of a new minimum norm solution, with more and better tuned parameters. This algorithm is now similar to the one implemented in MNE software.
Januray 2010
Project sources on default anatomy
Any source file estimated for a given surface can be re-projected on another surface. This allows to project sources of individual subjects on the default anatomy to compare subjects. See tutorial: Project sources on default anatomy.
Display channels in columns
The time series can be displayed in columns. The displayed sensors can be edited with an interface similar to the selection editor in MNE software.
December 2009
Automatic updates
Brainstorm is now self-updating. It tests at each startup if your version is older than a month; if so, it downloads a new version. The software can also be updated manually very easily (menu Help > Update Brainstorm).