Hello
I've got some confusing results which I was hoping someone might be able to shine a light on for me.
I've been looking at comparing different conditions, both in terms of sensor data and source data. I have 8 conditions, up to 4 trials each.
For the sensor data, I computed PSDs on the trials in frequency bands, and then averaged these for each participant (Average everything -> arithmetic average, with the "exclude flat signals from the average checked). I then normalised the average PSD file through spectrum normalisation -> relative power (because this is what is advised for the source data, so I wanted to be consistent).
For the between participant analysis I then either performed a permutation t-test on the averaged PSD files from each participant e.g. t-test(PSD(trialsA), PSD(trialsB)).
For comparison, I also ran a difference of the averaged trials A and trials B from all participants, as per the average(PSD(trialsA)) - average(PSD(trialsB)) instruction in this post: What should be compared in statisctics? - #3 by soltanlou. In other words, for each condition, I averaged the participants' average PSD files to get a group mean.
I was expecting to find the same results, but with different scale bars i.e. t-statistic vs. no units, but I see quite different results for all the frequency bands.
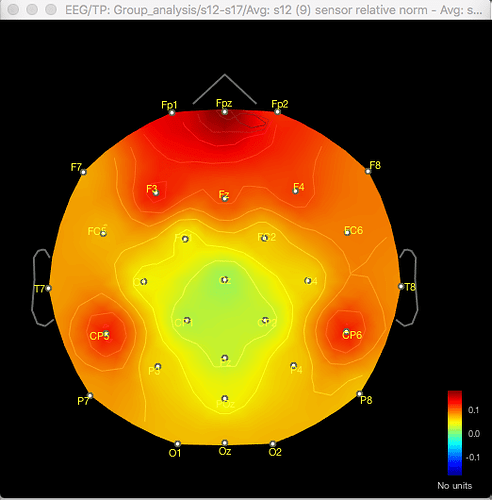
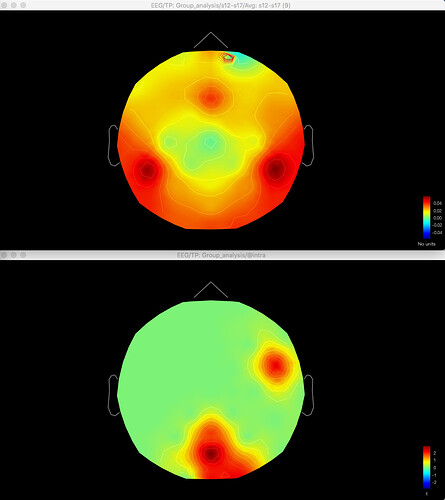
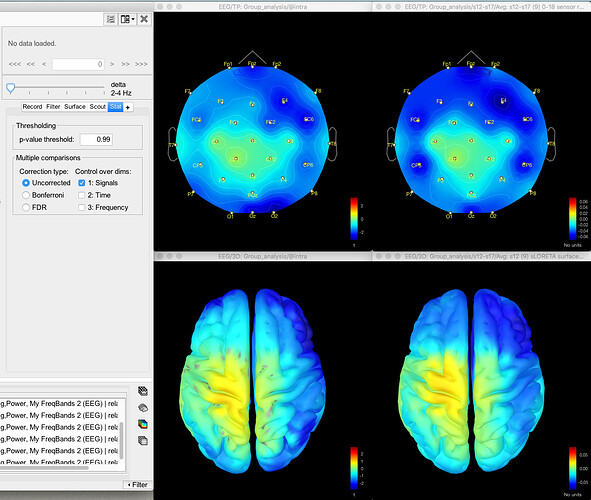
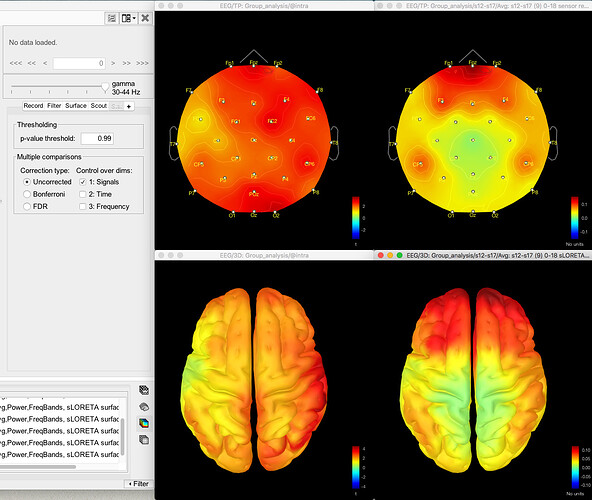
Here's the difference 2D sensor map e.g. for gamma for one contrast:
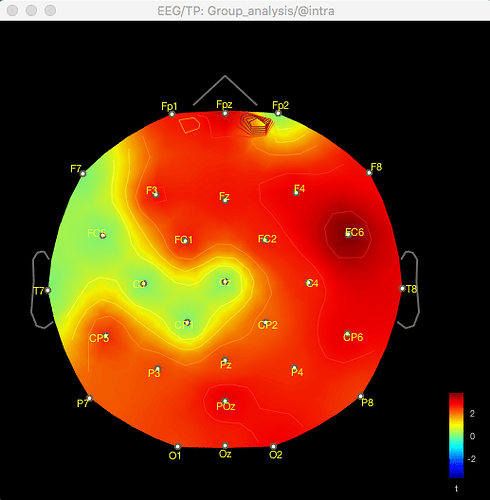
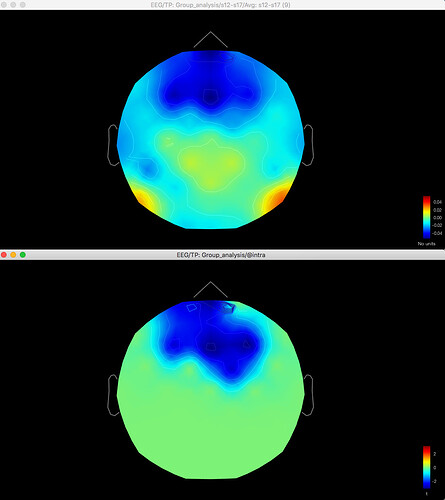
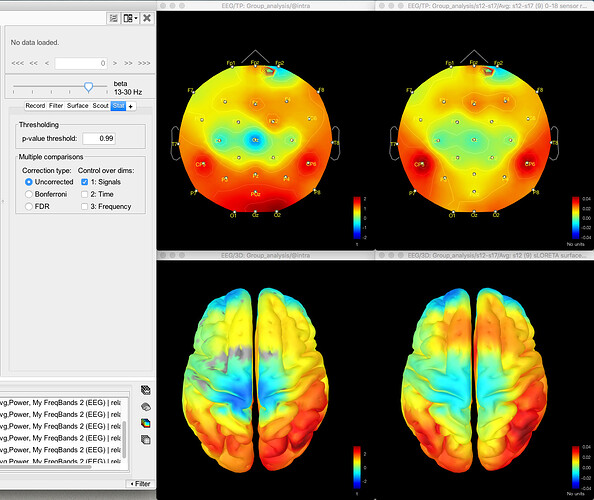
And here's the result of the t-test for the same data for gamma (no multiple corrections applied):
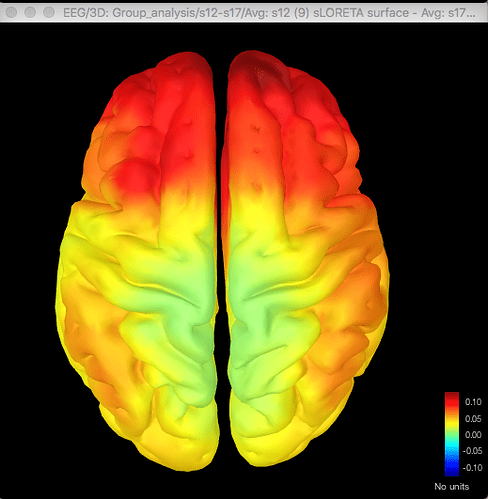
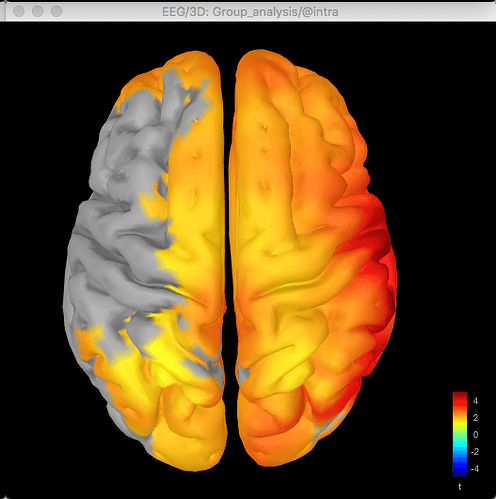
The same discrepancy is seen when I use the same approach for looking at the unconstrained sLORETA surface model source data (on averaged PSD files for each condition from each participant, with the spectrum then normalised to the relative power) - the source data seems to match the distribution of the sensor data for the differences between conditions or for the t-test between conditions, but both give different results.
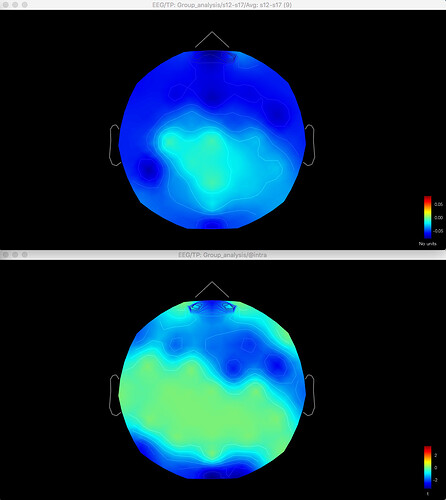
Here's the difference map of the source data, with the same data for gamma as shown above for the sensor data:
Here's the source data for the t-test (no multiple corrections applied):
So with the difference maps, the effect seems to be largely frontal and parietal (which is more consistent with my hypothesis), but with the t-test it is more lateralised to the right hemisphere, primarily in the temporal lobe.
When I export the relative power PSD values from the sensor data and run a repeated measured ANOVA in another software program on the different conditions, the significant results that come out match the areas of greater activation in the difference map from the sensor data, rather than the t-test map.
I wondered if it was because I was comparing a parametric test (ANOVA) with a non-parametric permutation test, so I ran a normal paired t-test on the data in Brainstorm, but that looked pretty much the same as the non-parametric test, and still different from the difference maps. So it wasn't that.
Am I missing something obvious?!
Many thanks
Luli