I am trying to import ECOG electrodes from curry but for some reason they are always in the wrong position and completely unrelated to the MRI that I importedto brainstorm. I am trying to import ECOG positions in MNI , PAN , and other coordinates. I have the NAS, RPA and LPApoints in my file.
What should be the type of coordinate*system for this to work? Why can't I just import the CT scan and mark the electrodes manually in brainstorm?
You can load the CT after loading the MRI into brainstorm and select the points within brainstorm. See my post here it may help:
http://neuroimage.usc.edu/forums/showthread.php?2645-SEEG-ECOG-electrode-locations
Thanks. I was able to import the CT but marking the electrodes using brainstorm was difficult. Now, I managed to import the electrode positions from Curry and I knew how to make them align beautifully with the MRI in brainstorm.
I want to ask you which method of source reconstruction in your opinion is best for ECoG?
The alignment MRI-CT and marking of the contacts has been improved recently:
http://neuroimage.usc.edu/brainstorm/News#CT-MRI_coregistration
If you have the NAS/LPA/RPA points in your file, it should be converted automatically to the coordinate system in Brainstorm.
About the source estimation method: you can expect the same kind of results as described in the source estimation tutorial:
http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation
In general, do not expect the sources extrapolated more than a few millimeters away from the contacts to be very meaningful. If the activity in a region was not recorded directly with an ECoG electrode, it cannot be reconstructed with this type of model.
Francois
How did you import the Curry electrode positions. From the .pom file?
I don’t know what .pom files are…
Usually it is easy to save the electrodes positions in simple ASCII text files.
Are you having problems with the file formats for the electrode positions?
I have done this a couple of times. I think I copied the coordinates of the electrodes from the .pom file to a plain txt file, with the channel numbers preceding the coordinates. Then copy and paste the NAS, RPA and LPA coordinates before the electrode coordinates.
Or also, I think what you can do is just import the CURRY functional data file right into brainstorm, in that file are the coordinates and ECOG data together. That is much easier.
I’m not at my CURRY workstation right now so I can’t confirm what I say, I may not be 100% correct.
Using only 3 fiducial points does not seem adequate. There were earlier posts about using surface points to better refine the fit. I'd like to do this but do people know how?
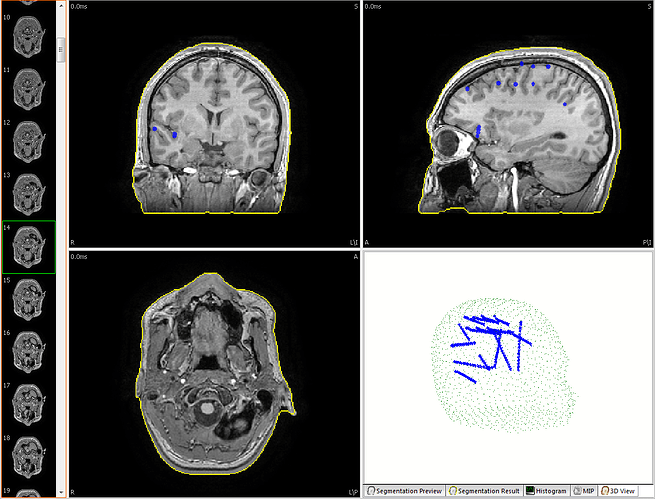
This is from Curry, showing electrodes tagged in blue surface points in green. Brainstorm should be able to fit the orientation by the surface points but I don't know the file format/structure this would need to be in to read it into Brainstorm.
Let me answer my own question - I figured it out.
In Curry, create ‘surface points’. I used a 10mm spacing which creates ~1400 points. Then import points to localize. Then export localize to matlab which creates a work space called ‘curryloc’.
You need to invert the matrix:
curryloc_alt = curryloc’
Then save this as an ASCII:
save (‘curryloc.txt’, ‘curryloc_alt’, ‘-ascii’)
This file can then be imported into Brainstorm as head points. It will query the need to scale and suggest 0.1.
Then refine using head points as usual.
It sounds like a good solution.
However, if you use these additional head points, I think your should NOT include the NAS/LPA/RPA points in the file with the coordinates of the ECOG contacts.
Otherwise, it would first use them to transform the coordinate system to SCS, then it would import the head points in this modified space and the result of the registration would be unpredictable.
It’s either the fiducials or the head points. Note that if the NAS/LPA/RPA points are placed precisely (same MRI slices in Brainstorm and Curry), the solution with the three fiducials is perfectly accurate.
Are the POM files simple text files with the contact coordinates and the head points?
If so, please send me an example file and I will add support for this file format in Brainstorm.
Thanks
Francois
I should add that you need to explicitly set the Curry coordinate to CTF so that it matches Brainstorm. I like having the extra surface points - and I don’t include the fiducials with it.
But I’ve also attached a .pom file (saved as .txt) so that you can see the organization. This is only for the fiducial points in the native Curry coordinate system (PAN). I never use this but it will be more helpful than arbitrary points.
Thank you for the file, I’ve added the support for the Curry .pom files in Brainstorm (update Brainstorm to get this last update).
However, I’m not sure they should be used in this way.
What you should import is a file that contains BOTH the position of the fiducials (NAS+LPA+RPA) and the position of the SEEG or ECOG contacts.
- The referential you use should not matter (you shouldn’t have to select “CTF” coordinates in Curry)
- The fiducials list should not include any additional point (remove all the others: Inion, AC, PC and IH)
Francois
Thanks for adding this - however I'm using the .pom file differently, only for surface points without fiducials. The reader seems to be looking for fiducials. I bring the fiducials is separately using the Curry .rs3 file (containing electode positions and fiducials). The .pom is only for additional surface points to better correct the co-registrations. Like MEG, only using 3 fiducials seems inadequate, and small errors have even more important consequences. John Mosher wrote earlier about doing it this way.
I had a work around above, so if I'm asking for too specific a change, don't bother with it.
Thanks.
Surface.pom (109.2 KB)
Hello
I fixed a bug that prevented reading the .pom head shape you sent.
Let me know if you think there is anything else that should be modified.
Francois
Surface.pom (109.2 KB)
Thank you for fixing this however the orientation doesn't read correctly. These surface points were created in Curry (PAN coordinate system). I've attached the .pom file.
It is normal that these head points are not in the same coordinates system. The point of these head points is to have them available in the same coordinates system as the sensors positions, so that you can register the sensors with the MRI.
http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Automatic_registration
To check the relative position of the head points and the ECOG contacts: right-click on the channel file > MRI registration > Check.
To run the automatic registration MRI-sensors based on the head points: Use the menu MRI registration > Refine using head points. Alternatively, right-click on the channel file > MRI registration > Edit, and use the equivalent button.
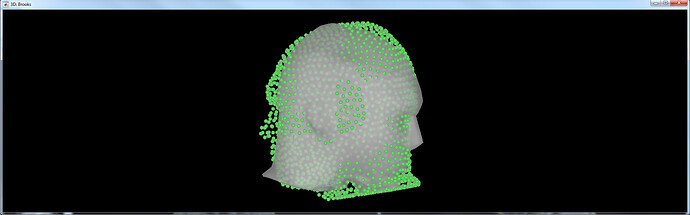
After running this automatic registration, is this a rotation issue (ie. the green points fit perfectly the grey surface), or is there a problem of flip left-right?
Thanks for the reply. I understand how the head points are to be used. I guess my trouble arises in that I have no way to build a file in Curry (.pom or otherwise) that contains both electrode positions with respect to fiducials and these extra surface points. I bring in the electrodes in the .rs3 file and then the head points in the .pom file.
Strangely, even after explicitly creating the .pom file using CTF coordinates in Curry the positions are still off in Brainstorm:
It looks like this is Curry:
Surface_CTF.pom (109.3 KB)
Of course if I 'refine using head points' in Brainstorm the electrodes will be shifted 90 degrees.
Now, I did work out a way of doing this - passing the Curry .pom out to Matlab and then reading it back into Brainstorm, so I can do what I need to do. I just bring it to you attention if there is a 'fix' to this, as others might want to use a similar workflow.
Thanks.
FYI: We improved significantly the tools available in Brainstorm for processing and visualizing SEEG and ECOG data, including new options for volume coregistration. They are now documented in a new tutorial:
http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
Hello,
I made a lot of progresses in importing SEEG contacts from various coordinates systems. It is now possible to import them in MRI “world” coordinates (the coordinates you see if you open the volumes in MRIcron, SPM or many other programs).
Reading again the various threads of the topic, I realize that the import sequence should have been the following:
- Link the recordings to the database
- Right-click on the channel file > Add EEG positions > Import from file: select a file with the positions of the contacts (Curry coordinates)
- Right-click on the channel file > Digitized head points > Add points: Select a file with the head shape in the same Curry coordinates => If you check the registration of the contacts with the anatomy: the contacts and head shape should be correctly registered, but both should not be correctly aligned on the head surface.
- Right-click on the channel file > MRI registration > Refine using head points => This should align the Curry head shape on the Brainstorm head surface, and then apply the same transformation to the contacts.
If there are still issues with loading SEEG or ECOG coordinates from Curry to Brainstorm, could you send me the files are you are trying to align? (MRI file + contact coordinates + head shape)
Thanks
Francois
Hi Francois,
When I want to import the .pom file, I get this in the command line:
BST> Warning: No channel information was read from the file.
However, I can open the file as text and see the contact names and locations.
Any suggestions?
Thanks