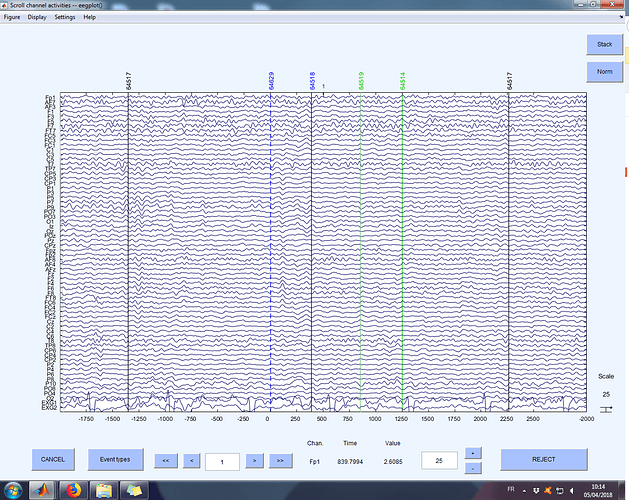
Hi Francois,
I am importing epoched data preprocessed in EEGlab. Everything works fine, I have the correct number of epochs (830 in my example subject, please find the file at this link), which have the proper length (from -2 to +3 s). As you can see from the 2 figures attached, the data are visually the same (I also checked the averages, they correspond).
The only problem is that all the markers look shifted in time in Brainstorm compared to EEGlab. This is the legend of the markers:
- 64517 = fixation
- 64629 = stimulus appearance
- 64518 = start response
- 64519 = end response
- 64514 = trial end
I imported the data both with the following code and manually (same result):
% start code
sublist= 2;
nsub = length(sublist);
for isub=1:nsub
isub
cat_dir= sprintf('myFolder' );
% create subject
SubjectName = sprintf('Subject%02d', sublist(isub));
UseDefaultAnat = 1;
UseDefaultChannel = 1;
[sSubject, iSubject] = db_add_subject(SubjectName, [], UseDefaultAnat, UseDefaultChannel);
% If an error occured in subject creation (subject already exists, impossible to create folders...)
if isempty(sSubject)
error('Could not create subject.');
end
sFiles = [];
% import EEG data
RawFiles = {...
strcat(cat_dir,sprintf('S%02d_final.set',sublist(isub)))};
sFiles = bst_process(...
'CallProcess', 'process_import_data_epoch', ...
sFiles, [], ...
'subjectname', SubjectName, ...
'condition', 'Alltrials', ...
'datafile', {{RawFiles{1}}, 'EEG-EEGLAB'}, ...
'iepochs', [], ...
'createcond', 0, ...
'channelalign', 1, ...
'usectfcomp', 1, ...
'usessp', 1, ...
'freq', [], ...
'baseline', []);
end
% end code
Is there a way I can fix this? Thank you very much for your help!
Rocco