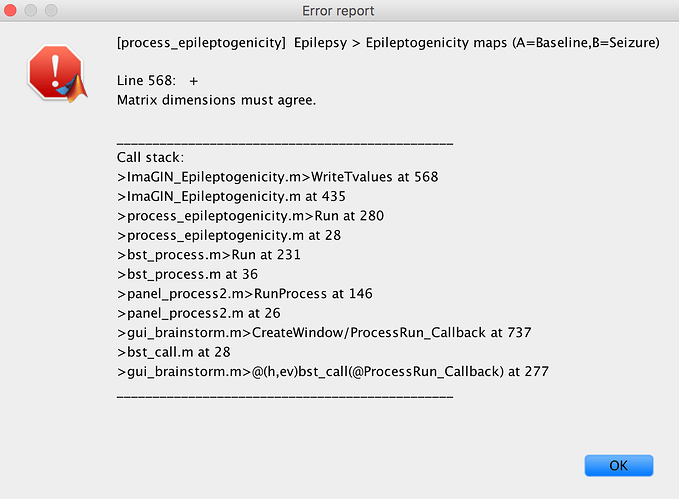
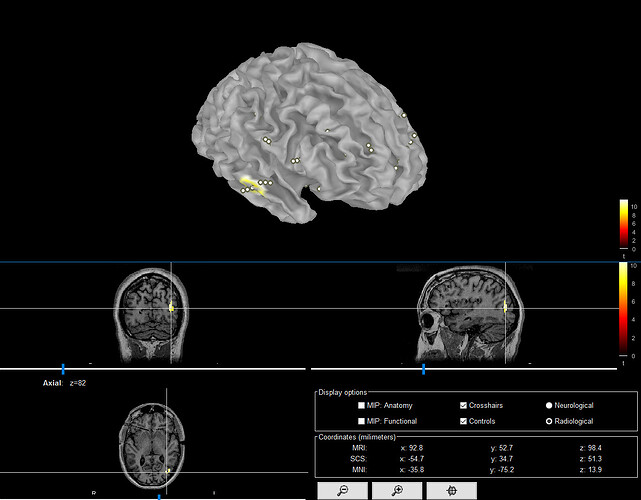
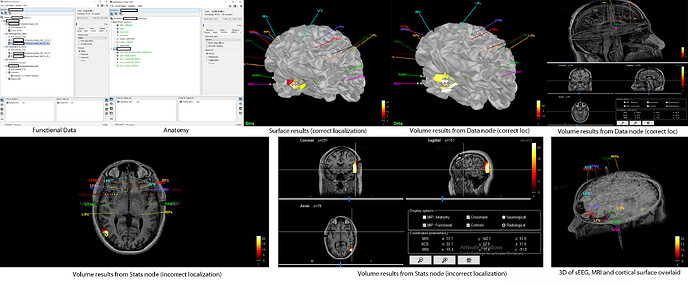
I am having a separate issue. I spent a lot of time importing preimplant MRI, postimplant CT, and making surfaces and locating electrodes manually. Then I try to do the Help>guidelines> epileptogenicity maps and it asks for MRI. I tried to do so and it gave an error and moreover deleted my previous surfaces and MRI. When I got to brainstorm db anatomy folder there is none of my work. Any way to recover it?
Thanks.
PS Matlab code preceding this is:
Error using load
Unable to find file or directory 'C:\Users\Cory
Myers\Documents\Duke\CNP2021\ResearchProjects\MatlabPackages\brainstorm_db\Protocol02\anat\07CM\subjectimage_2021-02-23_15-04_axspgr_0002_spm_reslice.mat'.
Error in in_mri (line 149)
MRI = load(MriFile);
Error in import_mri (line 148)
sMri = in_mri(MriFile, FileFormat, isInteractive && ~isMni, isNormalize);
Error in scenario_epilepto>ValidateImportAnatomy (line 346)
DbMriFilePre = import_mri(iSubject, MriFilePre, 'ALL', 0, 0);
Error in scenario_epilepto>@(c)ValidateImportAnatomy() (line 109)
ctrl.fcnValidate{i} = @(c)ValidateImportAnatomy();
Error in panel_guidelines>SwitchPanel (line 116)
[isValidated, errMsg] = ctrl.fcnValidate{iPanel}();
Error in panel_guidelines>@(h,ev)SwitchPanel('next') (line 56)
ctrl.jButtonNext = gui_component('button', jPanelControl, 'br', '>>', buttonFormat, [], @(h,ev)SwitchPanel('next'));
** Error: The pre-implantation MRI file you selected does not exist.
BST> Saving protocol "Protocol02"...
BST> Emptying temporary directory...
BST> Brainstorm stopped.
![]()