Dear Francois,
thank you for these changes! You are right, marking bad channels in spectra already works. This is also very helpful.
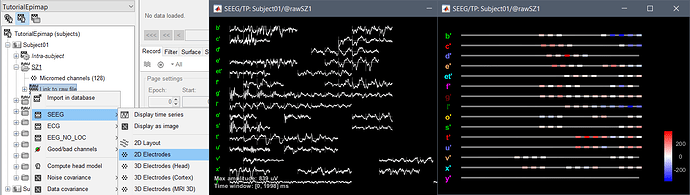
I very much like the new display of sEEG electrodes without 3D positions. I tried it on an ECoG dataset for which not all contact positions were marked yet and I get the following error. As I believe informations for these 2D plot are completely based on contact names, can you please enable 2D display without marked electrode positions?
Best regards
Marcel
BST> Saving scouts in surface: ECOG_tutorial/tess_cortex_pial_low.mat
BST> Warning: The position of the following sensors is not set: G_B1 G_B2 G_B3 G_B4 G_B5 G_B6 G_B7 G_B8 G_C1 G_C2 G_C3 G_C4 G_C5 G_C6 G_C7 G_C8 G_D1 G_D2 G_D3 G_D4 G_D5 G_D6 G_D7 G_D8 G_E1 G_E2 G_E3 G_E4 G_E5 G_E6 G_E7 G_E8 G_F1 G_F2 G_F3 G_F4 G_F5 G_F6 G_F7 G_F8 G_G1 G_G2 G_G3 G_G4 G_G5 G_G6 G_G7 G_G8 G_H1 G_H2 G_H3 G_H4 G_H5 G_H6 G_H7 G_H8 IHA1 IHA2 IHA3 IHA4 IHB2 IHB3 IHB4 IHC1 IHC2 IHC3 IHC4
Error using ./
Matrix dimensions must agree.
Error in panel_ieeg>AlignContacts (line 2211)
NewLoc = ((sElectrodes(iElec).Loc(:,1) * w1 + sElectrodes(iElec).Loc(:,2) * w2) ./ (w1 + w2))';
Error in panel_ieeg (line 30)
eval(macro_method);
Error in figure_3d>PlotSensors3D (line 2667)
Channel = panel_ieeg('AlignContacts', iDS, iFig, 'default', sElectrodes, Channel);
Error in figure_3d (line 40)
eval(macro_method);
Error in figure_topo>CreateTopo3dElectrodes (line 1303)
[PlotHandles.hSurf, MarkersLocs2D] = figure_3d('PlotSensors3D', iDS, iFig, Channel, ChanLoc, TopoType);
Error in figure_topo>PlotFigure (line 496)
CreateTopo3dElectrodes(iDS, iFig, Channel(selChan), markers_loc(selChan,:), TopoInfo.TopoType);
Error in figure_topo (line 27)
eval(macro_method);
Error in view_topography (line 379)
isOk = figure_topo('PlotFigure', iDS, iFig, 1);
Error in tree_callbacks>@(h,ev)view_topography(FileName,Modality,'2DElectrodes') (line 2658)
gui_component('MenuItem', jMenu, [], '2D Electrodes', IconLoader.ICON_CHANNEL, [], @(h,ev)view_topography(FileName, Modality,
'2DElectrodes'));