Hello,
I am a newbie to Brainstorm and source localisation. I have a long continuous EEG recording (Epileptic), nearly an hour. I wanted to source localise using this data. I built mne-sLORETA model. I am encountering few problems. Mu ultimate aim is to get times series for parcellated regions (68 regions that I parcellated cortex using freesurfer).
-
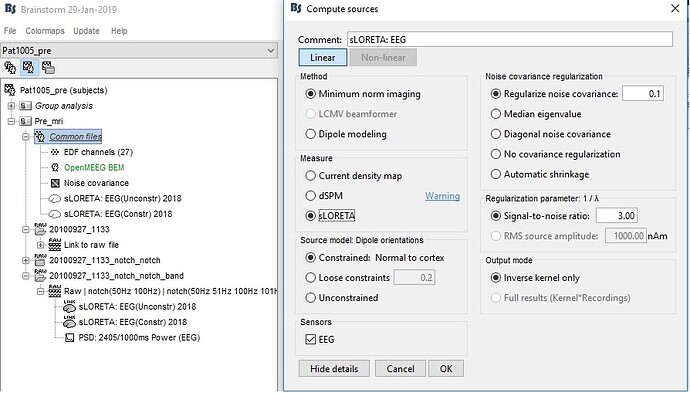
I would like to have source time series for 15000 sources. I searched the forum and found one relevant discussion. However, I am unable to reproduce what Francosis has suggested there as in "Output Mode" only "Inverse Kernel" gets highlighted ("Full Results" cannot be selected). May I know why is this happening?
-
I also tried the second procedure (in_bst_results) suggested by Francosis, even this one did not work. Unfortunately, I couldn't see the EEG time series at all when I imported into MATLAB (also checked in database folder, the filtered file is only 7KB). Can you please help me with this?
-
My ultimate aim is to get time series for 68 parcellated (parcellated) FreeSurfer. I down-sampled 15002 vertices to 68 regions-Desikan-Killiany (Scouts/ROIs). Even here, I tried to export/save scout time series but couldnt find the matrix (68xtime points) either in MATLAB workspace or on hard-drive.
In case these questions were answered earlier, please accept my apologies and point me to those. Any help is much appreciated.
Regards,
Sri.