Hi,
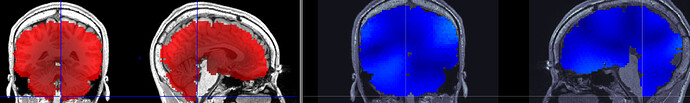
I've computed the PSD for my source space data and have successfully exported as a volume (SPM8). However when I look at the data overlayed on the same MRI in mricron, it looks like it's off a bit (see image). Red is data exported as 3D in mricron. Blue is the same data displayed in Brainstorm. I put both cursors at the same coordinates. I've triple checked that it's the same subject.
Is there something I'm missing?
Thanks,
Elizabeth
Also, I forgot to mention that the two viewers have a R/L flip when viewing. The data is the same. I’m worried that the data looks like it’s shifting down/inferior.
Hi Elizabeth,
Have you processed the MRI with FreeSurfer? Are your sources in Brainstorm estimated from the FreeSurfer anatomy? And the volume you used in mricron is the original .nii you passed to FreeSurfer for segmentation?
If so, it is normal not to have the two volumes aligned: FreeSurfer rewrites the volume in its own referential without keeping the initial .nii “voxel to world” transformation (the file T1.mgz, imported by Brainstorm). There is maybe a way to read it from some of the FreeSurfer outputs, but I don’t know how.
If you export the MRI from Brainstorm as .nii (go to the subject’s anatomy, right-click on the MRI > File > Export to file), and overlay the PSD results on it, they should align correctly.
Let me know if the problem is not the one I mentioned. There are probably other issues related with these world coordinates in the .nii files. If you imported the files recently, the sform/qform matrices might not have been imported correctly. This code was heavily edited during the summer and fall 2017.
Cheers,
Francois
Thanks Francois!
If I export the .nii from Brainstorm, the PSD results do align nicely. However, it’s not overlaying on the T1.mgz from the freesurfer folder that was loaded into Brainstorm. It seems like somewhere between loading it into Brainstorm and exporting it from brainstorm, it gets shifted. I thought maybe this was just a case of the .mgz to .nii conversion. However, when I convert the T1.mgz to T1.nii using the mri_convert command, it still doesn’t overlay properly.
I think the worse-case scenario here is to export the T1 from brainstorm and register this to the subject’s original T1, but I’d like to avoid that step if I can.
I realized that I could easily get the original transformation voxel=>world from the .mgz files. I tried to add this in the code but could not test it much (update brainstorm to get this fix).
This transformation will be used when exporting .nii volumes from Brainstorm, and hopefully this exported volumes will align with the T1.mgz file converted to T1.nii with mri_convert.
The problem for your current database is that this transformation can only be loaded when importing the anatomy. You would have to start over completely from the import of the anatomy before being able to export more correct .nii files.
Otherwise, you could try making a copy of your subject (right-click > File > Duplicate subject), then right-click on the subject > Import anatomy folder again. At that point, you would need to select EXACTLY the same NAS/LPA/RPA points as previously (you could use the popup menu of the MRI viewer to copy the fiducials from the old subject, and paste them when importing the new one).
This is a bit complicated, so if you need additional explanations, don’t hesitate to ask again.
In any case, could you please send me the two files T1.mgz and T1.nii you have? I would like to test if it is equivalent to importing the volume T1.mgz in Brainstorm and exporting it as .nii.
You could zip and upload these files somewhere, then send me the link as a private message on this forum.
Thanks
Francois