Tutorial 28: Connectivity
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Hossein Shahabi, Raymundo Cassani, Takfarinas Medani
Cognitive and perceptual functions are the result of coordinated activity of functionally specialized regions in the brain. Brain connectivity investigates how these different regions (or nodes) interact as a network, with the goal of having a better understanding of how the brain processes information. Depending on which connectivity characteristic is studied, a distinction is made between structural (fiber pathways), functional (non-directed statistical dependency) and effective (causal interaction) connectivity between regions. Effective connectivity is often referred as directed functional connectivity. In this tutorial we will see how to compute different connectivity metrics for non-directed and directed functional analyses in Brainstorm, first with simulated data and later with real data.
Contents
- General considerations in connectivity analysis
- Simulated data (MAR model)
- Correlation
- Coherence
- Granger Causality
- Spectral Granger causality
- Envelope Correlation (2020)
- Phase locking value
- Method selection and comparison
- Sensor- and source-domain connectivity analyses
- Connectivity measure on real data : MEG/EEG data
- TODO : Connectivity measure on real data : MEG/EEG data
- Sections to add
- On the hard drive
- Additional documentation
General considerations in connectivity analysis
Connectivity analyses are commonly performed by computing a bivariate connectivity metric for all the possible pairs of time series or signals. The result of such approach can be presented as a connectivity graph (left image), where each signal is represented as a node, and the value of the connectivity metric is the value of the edge between the corresponding nodes. This graph representation becomes overwhelming when too many nodes are considered, as such, the connectivity graph can be represented with its connectivity matrix, aka adjacency matrix (right image).
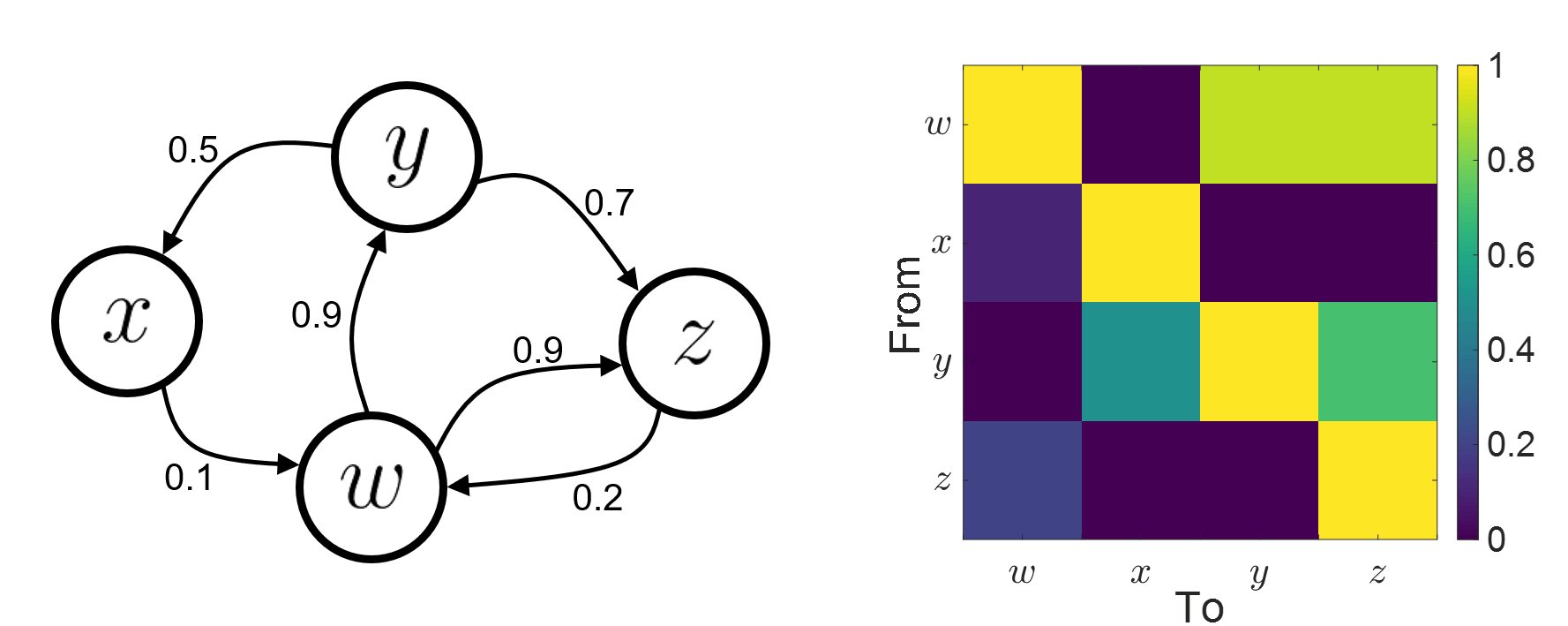
Sensors or sources: The signals used for the connectivity analysis can be derived from the sensor data (EEG/MEG signals) or from the reconstructed sources (voxels or scouts).
Directed and non-directed: The direction of the interaction between signals (as statistical causation) can be measured with directed metrics. However, this is not possible with non-directed metrics, as result, the connectivity metric "from Signal  to Signal
to Signal 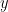 " is equal to the connectivity metric "from Signal
" is equal to the connectivity metric "from Signal 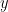 to Signal
to Signal  ".
".
Recording condition: While connectivity analysis can be performed on resting-state (spontaneous) and event-related (trials) recordings, the appropriate connectivity method depends on the recording condition.
Full network vs point-based connectivity: In full network, the connectivity metric is computed for all the possible node pairs in the network (N×N approach), and gives as result a detailed connectivity graph. Alternatively, the analysis can be performed solely between one node (aka seed) and the rest of the nodes in the network (1×N approach), this approach is faster to compute and is more useful when you are interested in the connectivity of a specific sensor or source.
Temporal resolution: Connectivity analyses can be performed in two ways: static and dynamic. Time-varying networks can present the dynamics of brain networks. In contrast, the static graphs illustrate a general perspective of brain connectivity which is helpful in specific conditions. Users need to decide which type of network is more informative for their study.
Time-frequency transformation: Several connectivity metrics rely on the time-frequency representation of the data, which is obtained with approaches such as the short time Fourier transform, Hilbert transform, and Morlet wavelet.
Simulated data (MAR model)
To compare different connectivity metrics, we use simulated data with known ground truth. Consider three channels constructed using the following multivariate autoregressive (MAR) process of 4th order.

where  with
with 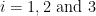 are coefficients of 4th order all-pole filters.
are coefficients of 4th order all-pole filters.
To compute these coefficients, we can consider a frequency response with desired pole and zero locations and use MATLAB zp2tf function for finding them.
Here, these coefficients were calculated in a way that the first channel has a dominant peak in the beta band (25 Hz), the second channel shows the highest power in the alpha band (10 Hz), and the third channel a similar level of energy in both bands. Additionally, the signal in the third channel is influenced by the signal in the first channel by the filter  .
.
We simulate data using the ARfit process. To run that, first clear the process panel and then select simulate -> simulate AR signals (ARfit) and use the following coefficients
box for code Screenshot
For a MAR model, the transfer function (or frequency response) is defined as:
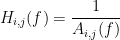
The diagonal elements show the auto-transfer function, which in our specific case is the spectrum of the signals. The off-diagonal terms represent the interactions between different signals. Here, we see the transfer function from channel 1 to channel 3. These transfer functions are our ground truth for connectivity values.
In the next sections we will compute different connectivity metrics for these simulated signals. As such, place the simulated data in the Process1 tab, select recordings, click on [Run] ( ![]() ) to open the Pipeline editor, and select the connectivity metric.
) to open the Pipeline editor, and select the connectivity metric.
Correlation
Correlation is a non-directed connectivity metric that can be used to show similarity, dependence or association among two random variables or signals. While this metric has been widely used in electrophysiology, it should not be considered the best technique to evaluate connectivity. The correlation metric by its nature fails to alleviate the problem of volume conduction and cannot explain the association in different frequency bands. However, it still can provide valuable information in case we deal with a few narrow-banded signals.
Let's compute the correlation for the simulated signals. Select the Connectivity » Correlation NxN process.
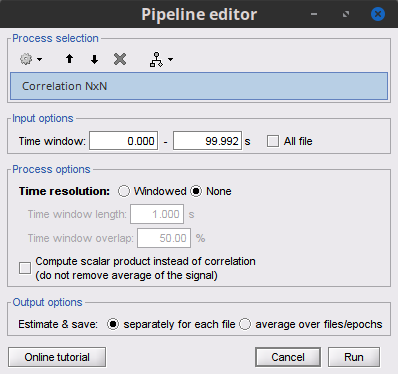
Process options
Time window: Segment of the signal used for the connectivity analysis. Check All file.
Sensor types or names: Leave it empty.
Include bad channels: Check it.
Process options: Uncheck it so the mean of the signals will be subtracted before computing the correlation.
Output configuration: Select Save individual results.
Result visualization
After running a N×N connectivity process, the results are stored as a N×N connectivity file (with the icon ![]() ). Right-click on this file to see its display options:
). Right-click on this file to see its display options:
Display as graph: plots the connectivity graph using a chord diagram where the color of the edges shows the connectivity metric value.
Display as image: plots the adjacent matrix for the connectivity file.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
In Display as image, the value of the connectivity metric between a signal and itself plotted as zero so that it doesn't force scaling the colormap to 1 if the other values are much smaller.
Coherence
Coherency or complex coherence, 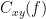 , is a complex-valued metric that measures of the linear relationship of two signals in the frequency domain. And, its magnitude square coherence (MSC),
, is a complex-valued metric that measures of the linear relationship of two signals in the frequency domain. And, its magnitude square coherence (MSC),  , often referred to as coherence, measures the covariance of two signals in the frequency domain. For a pair of signals
, often referred to as coherence, measures the covariance of two signals in the frequency domain. For a pair of signals 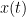 and
and 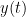 , with spectra
, with spectra 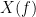 and
and 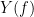 , the MSC is defined as:
, the MSC is defined as:
Two related measures, which alleviate the problem of volume conduction, are ?imaginary coherence, 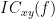 , and the ?lagged coherence,
, and the ?lagged coherence, 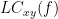 , which are defined as:
, which are defined as:
where 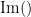 and
and 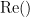 describe the imaginary and real parts of a complex number.
describe the imaginary and real parts of a complex number.
To calculate coherence values in Brainstorm, select the Connectivity » Coherence NxN process.
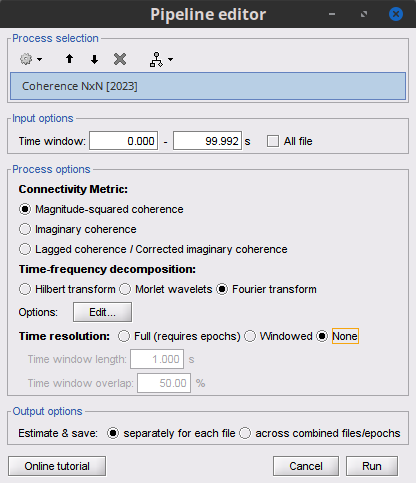
Process options
Time window: Segment of the signal used for the connectivity analysis. Check All file.
Sensor types or names: Leave it empty.
Include bad channels: Check it.
Removing evoked response: Check this box to remove the averaged evoked
Process options: Different types of coherence. Select Magnitude squared coherence.
Overlap for PSD estimation: Percentage of overlap between consecutive windows for PSD estimation.
Maximum frequency resolution: Width of frequency bins in PSD estimation.
Highest frequency of interest: Highest frequency for the analysis, it should be <= Fs/2.
Output configuration: Select one file per input file.
Result visualization
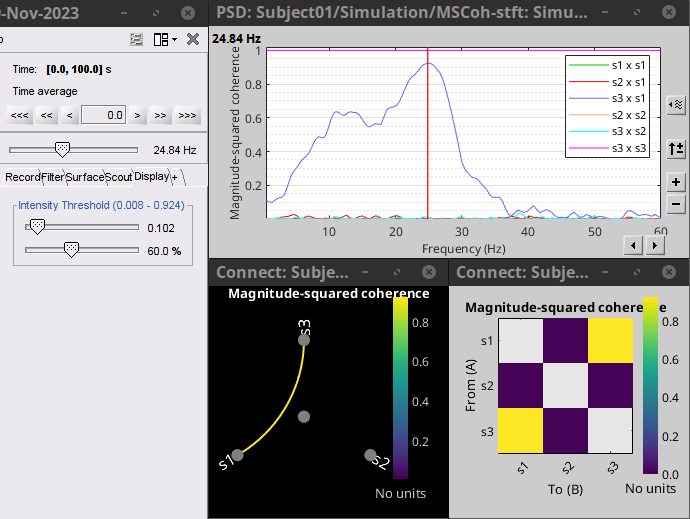
Coherence is a function of frequency, as such, for each frequency point there is a connectivity graph and a connectivity matrix. Right-click on the coherence result file to see its display options:
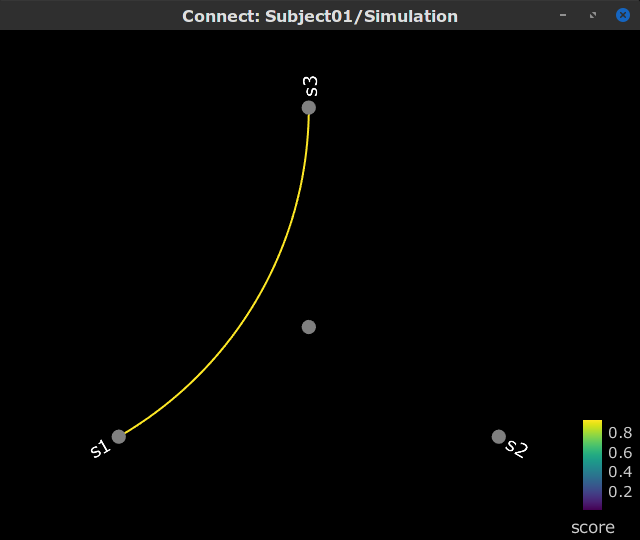
Display as graph: Plot the connectivity graph at a given frequency point.
Display as image: Plot the connectivity matrix at a given frequency point.
Power spectrum: Plot coherence as a function of frequency for all the possible node pairs.
By clicking on the spectral representation of the coherence we change the frequency that is displayed in the connectivity graph and matrix. This frequency can be also changed in the Time window.
In the same way, we can compute the other types of coherence. The figure below presents the spectra for the imaginary coherence (left) and the lagged coherence (right).
We see the last two measures are similar but have different values in several frequencies. However, both imaginary and lagged coherence are more accurate than coherence.
Granger Causality
Granger causality (GC) is a method of directed functional connectivity, which is base on the Wiener-Granger causality methodology. GC is a measure of linear dependence, which tests whether the prediction of signal A (using a linear autoregressive model) is improved by adding signal B (also using a linear autoregressive model). If this is true, signal B has a Granger causal effect on the first signal A. In other words, independent information of the past of signal B improves the prediction of signal A obtained with the past of signal A alone. GC is nonnegative, and zero when there is no Granger causality. As only the past of the signals is considered, the GC metric is directional. The term independent is emphasized because it creates some interesting properties for GC, such as, that it is invariant under rescaling of A and B, as well as the addition of a multiple of A to B. See Granger causality - mathematical background for a complete formulation of the method.
Despite the name, Granger causality indicates directionality but not true causality.
For example, if a variable C is causing both A and B, but with a smaller delay for B than for A, then a GC measure between A and B would show a non-zero GC for B->A, even though B is not truly causing A (Bressler and Seth, 2011).
To compute the Granger causality values in Brainstorm, select the Connectivity » Bivariate Granger causality NxN process.
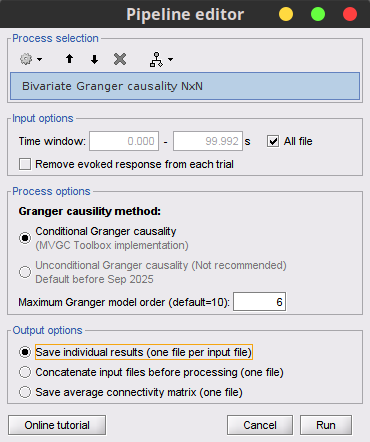
Process options
Time window: Segment of the signal used for the connectivity analysis. Check All file.
Sensor types or names: Leave it empty.
Include bad channels: Check it.
Removing evoked response: Check this box to remove the averaged evoked. It is also recommended by some as it meets the zero-mean stationarity requirement (improves stationarity of the system). However, the problem with this approach is that it does not account for trial-to-trial variability (For a discussion see (Wang et al., 2008) todo-> link to paper).
Model order: The most common criteria used to define the order of the model are the ?Akaike’s information criterion, the ?Bayesian-Schwartz’s criterion, and the ?Hannan-Quinn criterion. Too low orders may lack the necessary details, while too big orders tend to create spurious values of connectivity. While our simulated signals were created with a model of 4, here we used as model order of 6 for a decent connectivity result.
Save individual results (one file per input file): option to save GC estimates on several files separately.
Concatenate input files before processing (one file): option to save GC estimates on several files as one concatenated matrix.
Result visualization
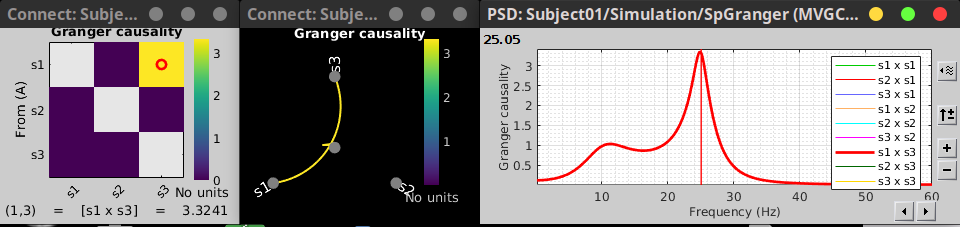
The GC results with the simulates signals are below. As GC metric is not symmetric, the connectivity matrix (right) is not symmetric. The upper right element of this matrix shows there is a signal flow from channel 1 to channel 3. In the connectivity graph the directionality is shown as GRADIENT (TO BE UPDATED WITH THE NEW GRAPH LIBRARY).
Spectral Granger causality
GC lacks of resolution in the frequency domain, as such, spectral Granger causality was introduced in [REF]. This metric is found in Connectivity » Bivariate Granger causality NxN.
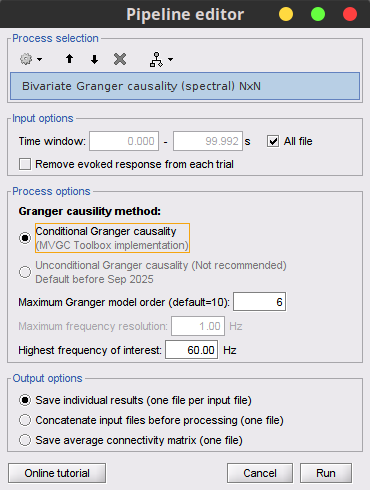
Process options
With respect to GC, spectral GC presents two extra parameters:
Maximum frequency resolution: Width of frequency bins in PSD estimation.
Highest frequency of interest: Highest frequency for the analysis, it should be <= Fs/2.
Result visualization
As with coherence, spectral GC can be plotted as a function of frequency. The plot below clearly shows a peak at 25 Hz, as expected.
Envelope Correlation (2020)
In the time-frequency tutorial the Morlet wavelets and Hilbert transform were introduced methods to decompose signals in the time-frequency (TF) domain. The result in this TF transformation can be seen as a set of ?analytic signals associated with narrowband (defined by the TF transformation method) signals. The analytic signal is a complex temporal representation of a real signal that has been useful in signal processing due to its characteristics, more specifically, its module and phase correspond to the instantaneous amplitude (or envelope) and instantaneous phase of the associated real signal.
The instantaneous amplitude (or envelope) of these band analytic signals can be used to carry out the pairwise connectivity analysis with metrics such as correlation and coherence (including lagged coherence). In computing the envelope correlation, an optional step is to orthogonalize the envelopes by removing their real part of coherence before the correlation. This orthogonalization process alleviates the effect of volume conduction in MEG/EEG signals. These connectivity metrics can be computed with the Connectivity » Envelope Correlation N×N [2020] process.
Process options
Time window: Segment of the signal used for the connectivity analysis. Check All file.
Sensor types or names: Leave it empty.
Include bad channels: Check it.
Removing evoked response: Check this box to remove the averaged evoked
Time-frequency transformation method: Either Hilbert transform or Morlet wavelets. Each of this methods requires additional parameters that are found in an external panel that opens by clicking on Edit. See the time-frequency tutorial
Signal splitting: This process has the capability of splitting the input data into several blocks for performing time-frequency transformation, and then merging them to build a single file. This feature helps to save a huge amount of memory and, at the same time, avoid breaking a long-time recording to short-time signals, which makes inconsistency in dynamic network representation of spontaneous data. The maximum number of blocks which can be specified is 20.
Connectivity measure: This is the connectivity metric that will be used with the envelopes.
Parallel processing: Enables the use of the parallel processing toolbox in Matlab to accelerate the computational procedure.
Output configuration: Generally, the above calculation results in a 4-D matrix, where dimensions represent channels (1st and 2nd dimensions), time points (3rd dimension), and frequency (4th dimension). In the case that we analyze event-related data, we have also several files (trials). However, due to the poor signal-to-noise ratio of a single trial, an individual realization of connectivity matrices for each of them is not in our interests. Consequently, we need to average connectivity matrices among all trials of a specific event. The second option of this part performs this averaging.
Phase locking value
An alternative class of connectivity metrics considers only the relative instantaneous phase between the two signals, i.e., phase-locking or synchronization (Tass et al., 1998). Phase locking is a fundamental concept in dynamical systems that has been used in control systems (the phase-locked loop) and in the analysis of nonlinear, chaotic and non-stationary systems. Since the brain is a nonlinear dynamical system, phase locking is an appropriate approach to quantifying connectivity. A more pragmatic argument for its use in studies of LFPs, EEG, and MEG is that it is robust to fluctuations in amplitude that may contain less information about interactions than does the relative phase (Lachaux et al., 1999; Mormann et al., 2000).
The most commonly used phase connectivity metric is the phase-locking value (PLV), which is defined as the length of the average vector of many unit vectors whose phase angle corresponds to the phase difference between two signals (Lachaux et al., 1999; Mormann et al., 2000). If the distribution of the phase difference between the two signals is uniform, the length of such an average vector will be zero. Conversely, if the phases of the two signals are strongly coupled, the length of the average vector will approach unity. For event-related studies, we would expect the marginal to be uniform across trials unless the phase is locked to a stimulus. In that case, we may have nonuniform marginals which could in principle lead to false indications of phase locking. between two signals.
PHASE TIME SERIES AND ROSE PLOT
Considering a pair of narrow-band analytic signals hats1(t) and hats2(t), obtained from the FT transformation using the Hilbert transform:
![\begin{eqnarray*}
PLV(t)= \left | E\left [ e^{j\Delta \phi (t)} \right ] \right | \\
\end{eqnarray*} \begin{eqnarray*}
PLV(t)= \left | E\left [ e^{j\Delta \phi (t)} \right ] \right | \\
\end{eqnarray*}](/brainstorm/Tutorials/Connectivity?action=AttachFile&do=get&target=latex_10f68832e53b9fba0f2c3add3817f05fdd0229f1_p1.png)
with:
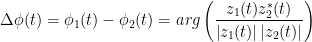
Process options
Time window: Segment of the signal used for the connectivity analysis. Check All file.
Sensor types or names: Leave it empty.
Include bad channels: Check it.
Frequency bands for the Hilbert transform: Frequency bands used for the time-frequency transformation with the Hilbert tranform method. See the time-frequency tutorial
Keep time information: Computes PLV across trials, thus the result is a PLV time series for each frequency band.
Measure: How the resulting vector is reported. Select Magnitude.
Result visualization
IMAGE
Method selection and comparison
We can have a comparison between different connectivity functions. The following table briefly does this job.
Comparing different approaches with the ground truth we find out that the HCorr function works slightly better than other coherence functions.
Sensor- and source-domain connectivity analyses
As mentioned above, the different connectivity metrics can be computed with sensor or source data. However, sensor-domain connectivity analyses present two important limitations:
They are not interpretable, as the relation between the estimated connectivity and the underlying neuroanatomy is not straightforward.
Sensor data is severely corrupted by effects of field spread and volume conduction. Due to these effects the activity of a single brain area could cause a spurious connectivity between MEG/EEG sensors.
Despite these limitations, sensor-domain connectivity analysis is commonly used. One approach to reduce the negative impact of field spread on connectivity analyses is to perform them in the source domain. In addition, source-domain connectivity analyses are interpretable as neuroanatomy is considered. As consequence, findings in this domain can be easily used in group studies using a normalization / registration. Regardless of the domain, it is highly recommended to have a clear hypothesis to test before starting the connectivity analysis. Although sensor- and source-domain connectivity analyses use different assumptions, the outcomes regarding the topology of the underlying networks should be consistent [Lai 2018].
Depending on the source estimation method the number of obtained sources can be in the order of tens of thousands, making the full connectivity analyses (N×N) impractical. As such, brain sources are often grouped in regions of intrests ROIs (scouts in Brainstorm). The most critical step in performing a source-domain connectivity analysis is the definition of ROIs, this is not a trivial procedure as it depends on the source estimation method, experimental task, and data available [Schhoffen 2009]. Common approaches found in the literature to select the ROIs for connectivity analysis are:
A prior: ROIs are selected based on a priori knowledge in a given experimental task
Coherence with a no-neuronal signal: ROIs that present a strong coherence with an external no-neuronal signal are selected.
Power maps: ROIs showing the strongest activity in an experimental task or ROIs that present the strongest differences of activity between experimental conditions are selected.
Full brain: A set of ROIs covering the full brain (cortical parcellations) is selected. In this selection approach, there are not any a priori assumptions or hypotheses. Although it can be used for exploratory analyses, it should not be taken as default for connectivity analyses.
Being an exploratory analysis, the full-brain connectivity analysis can help to get a better understanding of the acquired data, and to develop hypothesis to test. However its outcomes should not be considered conclusive, as they may be the result of circular analysis. [Kriegeskorte 2009].
The optimal selection of ROIs to perform source-domain connectivity analysis is still an open question.
Connectivity measure on real data : MEG/EEG data
Let's go back to our auditory oddball dataset. According to the literature, we expect to observe at least the following effects in their respective areas of the brain:
From 0 to 150 ms: we expect the bilateral activity in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
From 100 to 300 ms: Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
From 300 to 550 ms: Frontal regions activation related to the decision making and motorpreparation, for the deviant beeps only (after 300ms).
In the Scouts tutorial, we have created 4 scouts for these regions of intrestes: A1L and A1R for the left and right primary cortices, IFGL for the left inferior frontal gyrus, and M1L for the left primary motor cortex.
The first response peak (91ms), the sources with high amplitudes are located around the primary auditory cortex, bilaterally, which is what we are expecting for auditory stimulation.
For this experiment we will focus on the beta high and gamma frequency, as shown here https://brainworksneurotherapy.com/what-are-brainwaves, "Beta is a ‘fast’ activity, present when we are alert, attentive, engaged in problem-solving, judgment, decision making, or focused mental activity."
- 1 dataset:
S01_AEF_20131218_02_600Hz.ds: Run #2, 360s, 200 standard + 40 deviants
For this data we select three main time windows to compute the connectivity:
time 1 : 0-150 ms : we expect the bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
time 2 : 100-300 ms: Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
time 3 : 300-550 ms : Frontal regions activation related to the decision making and motorpreparation, for the deviant beeps only (after 300ms).
The computation are done here only for the second recording.
Connectivity is computed at the source points (dipole) or at a defined brain region also called scouts. The signal used art this level is obtained from the inverse problem process, in which each source-level node (dipole) is assigned with an activation value at each time point.
TODO : Connectivity measure on real data : MEG/EEG data
For all the brain imaging experiments, it is highly recommended to have a clear hypothesis to test before starting the analysis of the recordings. With this auditory oddball experiment, we would like to explore the temporal dynamics of the auditory network, the deviant detection, and the motor response. According to the literature, we expect to observe at least the following effects:
Bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
Decision making and motor preparation, for the deviant beeps only (after 300ms).
The first response peak (91ms), the sources with high amplitudes are located around the primary auditory cortex, bilaterally, which is what we are expecting for auditory stimulation.
For this experiment we will focus on the beta high and gamma frequency, as shown here https://brainworksneurotherapy.com/what-are-brainwaves, "Beta is a ‘fast’ activity, present when we are alert, attentive, engaged in problem-solving, judgment, decision making, or focused mental activity."
- 1 dataset:
S01_AEF_20131218_02_600Hz.ds: Run #2, 360s, 200 standard + 40 deviants
For this data we select three main time windows to compute the connectivity:
time 1 : 0-150 ms : we expect the bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
time 2 : 100-300 ms: Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
time 3 : 300-550 ms : Frontal regions activation related to the decision making and motorpreparation, for the deviant beeps only (after 300ms).
The computation are done here only for the second recording.
Sources level
Connectivity is computed at the source points (dipole) or at a defined brain region also called scouts. The signal used art this level is obtained from the inverse problem process, in which each source-level node (dipole) is assigned with an activation value at each time point.
Therefore, we can compute the connectivity matrix between all pairs of the node. This process is possible only of the inverse problem is computed (ref to tuto here).
To run this in brainstorm, you need to drag and drop the source files within the process1 tab, select the option 'source process' click on the Run button, then you can select the connectivity measure that you want to perform.
As in the previous section, we can compute the source connectivity matrix for each trail, then average overall trial. However, this process is time and memory consuming. For each trial, a matrix of 15002x15002 elements is computed and saved in the hard disc (~0.9 Go per trial). In the case of the unconstrained source, the size is 45006x45006.
This is obviously a very large number and it does not really make sense. Therefore, the strategy is to reduce the dimensionality of the source space and adopt a parcellation scheme, in other terms we will use the scouts. Instead, to compute the connectivity value values between two dipoles, we will use a set of dipoles pairs that belong to a given area in the cortex.
Although the choice of the optimal parcellation scheme for the source space is not easy. The optimal choice is to choose a parcellation based on anatomy, for example the Brodmann parcellation here. In brainstorm these atlases are imported in Brainstorm as scouts (cortical regions of interest), and saved directly in the surface files as explained in this tutorial here.
In this tutorial, we will use the scouts " Destrieux atlas" (following figure) ![]()
To select this atlas, from the connectivity menu, you have to check the box 'use scouts', select the scout function 'mean' and apply the function 'Before', save individual results.
In this tutorial, we select the correlation as example, the same process is expected for the other methods.
For more detail for these options please refer to this thread
The following matrix is the solution that we obtain with these scouts with the size of 148x148 for this atlas (~400 Ko)
For this data we select three main time windows to compute the connectivity:
time 1 : 0-150 ms : we expect the bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
time 2 : 100-300 ms : Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
time 3 : 300-550 ms : Frontal regions activation related to the decision making and motor preparation, for the deviant beeps only (after 300ms).
The computation are done here only for the second recording.
Coherence
Correlation
For this data we select three main time windows to compute the connectivity:
time 1 : 0-150 ms : we expect the bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
time 2 : 100-300 ms : Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
time 3 : 300-550 ms : Frontal regions activation related to the decision making and motor preparation, for the deviant beeps only (after 300ms).
The computation are done here only for the second recording.
For the time 1, We find high correlation value in both hemisphere on the temporal areas.
This connectivity is observed between the area 99 and 41 and between the 42 and 100 areas.
Corresponding to the name of the areas here
time 3 : 300-550 ms : Frontal regions activation related to the decision making and motor preparation, for the deviant beeps only (after 300ms).
The computation are done here only for the second recording.
Coherence
Correlation
For the time 1, We find high correlation value in both hemisphere on the temporal areas.
This connectivity is observed between the area 99 and 41 and between the 42 and 100 areas.
Corresponding to: name of the areas here
Similar results are observed either for the deviant and standard sounds.
For the time 3,
PLV
This connectivity is observed between the area 99 and 41 and between the 42 and 100 areas.
Corresponding to: name of the areas here
Similar results are observed either for the deviant and standard sounds.
For the time 3,
PLV
Using the option > right-click on figure> Graphic Options > Display Region max M or just use from the keyboard with M key.
TODO : Sensors level
Connectivity is computed at the sensors or the electrodes levels from the recorded time series.
PLV
Using the option > right-click on figure> Graphic Options > Display Region max M or just use from the keyboard with M key.
TODO : Sensors level
Connectivity is computed at the sensors or the electrodes levels from the recorded time series.
PLV
Using the option > right-click on figure> Graphic Options > Display Region max M or just use from the keyboard with M key.
TODO : discuss
- Explain or give more information about the methods and how to choose the best parameters
ex: plv better with 100 samples & narrow bands
Using the option > right-click on figure> Graphic Options > Display Region max M or just use from the keyboard with M key.
TODO : discuss
- Explain or give more information about the methods and how to choose the best parameters
ex : plv better with 100 samples & narrow bands
- Explain the choice either with ERP or without, and why (link to the cited paper, can't find it)
- Show/add other relevant measures of statistics to separate the two conditions
- Add the option : checkbox remove the erp for PLV and CORR and PTE
- ...
Sections to add
Discuss option concatenate vs. average for the estimation of coherence on single trials: https://neuroimage.usc.edu/forums/t/difference-between-averaging-coherence-and-concatenate/22726/4
- Discuss pValues computed for the various methods
On the hard drive
TODO: Document data storage.
Additional documentation
References
1. Reference #1
2. Reference #2
3. Reference #3
Articles
Lagged-Coherence: Pascual-Marqui RD. Coherence and phase synchronization: generalization to pairs of multivariate time series, and removal of zero-lag contributions, arXiv preprint arXiv:0706.1776. 2007 Jun 12.
Phase transfer entropy: Lobier M, Siebenhühner F, Palva S, Palva JM Phase transfer entropy: A novel phase-based measure for directed connectivity in networks coupled by oscillatory interactions, NeuroImage 2014, 85:853-872
Barzegaran E, Knyazeva MG. Functional connectivity analysis in EEG source space: The choice of method Ward LM, editor. PLOS ONE. 2017 Jul 20;12(7):e0181105.
Forum discussions
Connectivity matrix storage:http://neuroimage.usc.edu/forums/showthread.php?1796
Comparing coherence values: http://neuroimage.usc.edu/forums/showthread.php?1556
Reading NxN PLV matrix: http://neuroimage.usc.edu/forums/t/pte-how-is-the-connectivity-matrix-stored/4618/2
Export multiple PLV matrices: https://neuroimage.usc.edu/forums/t/export-multiple-plv-matrices/2014/4
Scout function and connectivity: http://neuroimage.usc.edu/forums/showthread.php?2843
Unconstrained sources and connectivity: http://neuroimage.usc.edu/forums/t/problem-with-surfaces-vs-volumes/3261
Digonal values: http://neuroimage.usc.edu/forums/t/choosing-scout-function-before-or-after/2454/2
Connectivity pipeline: https://neuroimage.usc.edu/forums/t/help-for-connectivity-pipeline/12558/4
Ongoing developments: https://neuroimage.usc.edu/forums/t/connectivity-tutorial-and-methods-development-on-bst/12223/3
Granger causality: https://neuroimage.usc.edu/forums/t/is-granger-causality-analysis-a-linear-operation/12506/12
Amplitude Envelope Correlation: * https://neuroimage.usc.edu/forums/t/problem-with-time-dynamic-envelope-correlation/21542/9

![\begin{eqnarray*}
IC_{xy}(f) = \frac{\mathrm{Im} \left (S_{xy}(f) \right )}{\sqrt{ S_{xx}(f)S_{yy}(f) }} \qquad \qquad \qquad \qquad LC_{xy}(f) = \frac{\mathrm{Im} \left (S_{xy}(f) \right )}{\sqrt{ S_{xx}(f)S_{yy}(f) - \left [ \mathrm{Re}\left ( S_{xy}(f) \right ) \right ]^{2} }} \\
\end{eqnarray*} \begin{eqnarray*}
IC_{xy}(f) = \frac{\mathrm{Im} \left (S_{xy}(f) \right )}{\sqrt{ S_{xx}(f)S_{yy}(f) }} \qquad \qquad \qquad \qquad LC_{xy}(f) = \frac{\mathrm{Im} \left (S_{xy}(f) \right )}{\sqrt{ S_{xx}(f)S_{yy}(f) - \left [ \mathrm{Re}\left ( S_{xy}(f) \right ) \right ]^{2} }} \\
\end{eqnarray*}](/brainstorm/Tutorials/Connectivity?action=AttachFile&do=get&target=latex_af67140a46334a4b50e13076d33d975708a33e91_p1.png)
This seems beyond the goal of the tutorial: Besides the original transfer functions, we can compute the directed transfer function (DTF) and partial directed coherence (PDC).![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)