Brainstorm's Database Structure
Authors: Martin Cousineau, Raymundo Cassani
Understanding how data is organized and managed in Brainstorm is paramount for scripting your own pipelines, and adding new processes in the software.
Conceptually, Brainstorm database consists in a collection of Protocols. In turn, a Protocol consists in a set of Subjects from whom Anatomical data (MRI) and recordings (MEG, EEG, fNIRS, SEEG, etc.) were acquired. Lastly, for each subject, the sensor locations, the recordings, the computed sources and all the data derived from their processing receive the name of Functional data.
This page describes the structure of the Brainstorm database on the hard drive, and the metadata that is necessary to manage it.
Contents
On the hard drive
Brainstorm database resides on the hard drive at the database directory (brainstorm_db/). The database consists in an organized collection of protocols, where each protocol each with its own directory, which has a strict hierarchy, explained in the bullet list below:

In anatomy data files and functional data files, the prefix in the filename indicates its content. For example, an anatomy file named surface_*.mat contains cortex surface information. Data in these files is stored as a Matlab structure, according to the different file types.
The content of the structure of each type_*.mat files is defined by the function db_template(), using type plus the mat suffix as argument. When the data contained in the type_*.mat file is loaded in memory in Brainstorm, it follows the Matlab structure given by db_template() using loaded plus the type. Lastly, the metadata related to the file type_*.mat is kept in memory as a Matlab structure given by db_template() using type as argument. For example:
db_template('surfacemat'): Structure saved on the hard drive as surface_*.mat file.
db_template('loadedsurface'): Structure saved in memory containing all the data loaded from the surface_*.mat file.
db_template('surface'): Structure saved in memory containing all the metadata for the surface_*.mat file, loaded from the protocol.db or protocol.mat files rather than the surface_*.mat file itself.
The protocol metadata files (protocol.mat and protocol.db) contain all the metadata that describe the relationship among the data files in the protocol directory. In case that any of the protocol metadata files is accidentally deleted or gets corrupted, the protocol metadata can be regenerated from all the brainstormsubject.mat and brainstormstudy.mat in the protocol directory as these files contain the metadata for the anatomy and functional data files for each subject in the protocol directory.
Protocol metadata
In Brainstorm versions <= 3.x, the protocol metadata was stored as a Matlab structure in the protocol.mat file, which content was kept in memory. Starting from version 4.0, the protocol metadata is stored as a SQLite database which is frequently queried, i.e., the protocol metadata is not kept in memory. Below a description of these two protocol metadata approaches.
Matlab structure: protocol.mat
Once a protocol is loaded, Brainstorm has Matlab structures in memory that contain the metadata of all the files in this protocol. These are located in the GlobalData.DataBase global variable, such that it is easily accessible in any function of the software. The structure of this variable is defined by db_template('GlobalData'). Let's walk through some of the important variables in this structure:
GlobalData.DataBase.ProtocolInfo: Basic information about the protocols in the Brainstorm database folder, refer to db_template('ProtocolInfo'). Fields:
Comment: Name of the protocol
STUDIES: Absolute path to the folder containing the functional data (/data/).
SUBJECTS: Absolute path to the folder containing the anatomical data (/anat/).
iStudy: Index of the currently selected study, if applicable.
UseDefaultAnat: Protocol setting, whether new subjects should use the default anatomy by default.
UseDefaultChannel: Protocol setting, whether new subjects should share channel files by default.
GlobalData.DataBase.ProtocolSubjects: Metadata of all subjects and their anatomical files of each loaded protocol, refer to db_template('ProtocolSubjects'). Fields:
Subject: Metadata of all regular subjects, refer to db_template('subject').
DefaultSubject: Special subject used to store default anatomy. Same structure as regular subjects.
GlobalData.DataBase.ProtocolStudies: Metadata of all studies and their functional files of each loaded protocol, refer to db_template('ProtocolStudies'). Fields:
Study: Metadata of all regular studies, refer to db_template('study').
DefaultStudy: Special study used to store Common files, e.g. a global channel file. Same structure as regular studies.
AnalysisStudy: Special study to store Inter-subject files, e.g. stats results. Same structure as regular studies.
GlobalData.DataBase.isProtocolLoaded: List of bool, whether the metadata of the protocol is already in memory or not. This is useful when you're running Brainstorm from the command line.
GlobalData.DataBase.isProtocolModified: List of bool, whether the metadata of the protocol was modified and therefore the protocol.mat file needs to be overwritten when Brainstorm closes.
GlobalData.DataBase.iProtocol: The index of the currently selected protocol.
GlobalData.DataBase.BrainstormDbDir: Absolute path to the brainstorm database directory.
GlobalData.DataBase.DbVersion: Version number of the currently loaded protocol. Used to update the protocol structure when new fields are added to a specific file for example. Refer to db_update().
GlobalData.DataBase.isReadOnly: Flag whether the user has write access to the protocol database directory. Used in tree_callbacks() to remove GUI option menus that would modify files.
GlobalData.DataBase.LastSavedTime: Timestamp of the last time the protocol.mat file was overwritten. There used to be an automatic save when it had been a few hours.
GlobalData.DataBase.Searches: Information about the search tabs of the database. Fields:
iActive: Index of the currently selected active search tab (0 if none).
Active: List of all active search tabs, with the first one being the whole database.
Name: Name of the tab, or empty if whole database
SearchNode: Tree structure representing the search query
AnatRootNode/FuncRootNode: The root node of the database tree, to avoid rebuilding the Java tree when you switch between tabs.
AnatSelNode/FuncSelNode: The currently selected node, to remember the last selected object before you switched between tabs.
All: If searches were saved between Brainstorm sessions, they are stored here.
Name: Name of the search
Search: Tree structure representing the search query
The two main variables of interest are ProtocolSubjects and ProtocolStudies. Indeed, together they contain the metadata of every single file in the protocol! Let's dive a bit deeper into both of them.
ProtocolSubjects This structure contains the metadata for the anatomy files of every subject. This metadata is organized in one structure per subject and has the following fields:
Name: Name of the subject. Two special subjects have specific names:
- @default_subject: Default anatomy subject.
- Group_analysis: Subject for statistical results or projections to a common space.
Comments: Additional information about the subject set manually by the user.
FileName: Path to the brainstormsubject.mat file where the subject specific metadata is stored in case the protocol's metadata file is corrupted or needs to be reloaded.
DateOfAcquisition: Unused, legacy field.
Anatomy: List of all MRI files, refer to db_template('Anatomy').
Surface: List of all surface and fibers files, refer to db_template('Surface').
iAnatomy: Index of the MRI file in the Anatomy list that is currently selected for this subject. This will show in green in the database viewer. Any operation related to this subject that requires an MRI file will load this selected one.
iScalp: Index of the selected scalp surface file from the Surface list.
iCortex: Index of the selected cortex surface file from the Surface list.
iInnerSkull: Index of the selected inner skull surface file from the Surface list.
iOuterSkull: Index of the selected outer skull surface file from the Surface list.
iFibers: Index of the selected fibers file from the Surface list.
iFEM: Index of the selected surface file used for FEM from the Surface list.
iOther: Index of the selected sub-structure surface file from the Surface list.
UseDefaultAnat: Whether this subject uses the default anatomy files rather than its own.
UseDefaultChannel: Whether this subject shares its channel file across all runs or uses the global channel file.
ProtocolStudies
This structure contains the metadata for the functional data files of every subject. This metadata is organized in one structure per study (also known as first level folder, or condition) and has the following fields:
Name: Name of the study. If it starts with "@raw", this is a special study that can only contain unprocessed files and is created when you create a new link to a raw file. Three special studies exist:
- @default_study: Common files shared either by the whole protocol (see @default_subject) or the whole subject. Typically channel files.
- @inter: Inter-subject statistical results.
- @intra: Intra-subject statistical results.
FileName: Path to the brainstormstudy.mat file where the study specific metadata is saved should the protocol metadata file get corrupted or need to be reloaded.
DateOfStudy: If this is a raw study, this is the acquisition date of the raw file. Otherwise, this is the date this study was created by the user.
BrainstormSubject: Path to the file of the metadata of the subject this study belongs to.
Condition: Either a duplicate of the Name field, or a cell array representing a sub-folder structure if this is a "sub-study", e.g. Subject01/ParentStudy/ChildStudy -> Condition = {"ParentStudy", "ChildStudy"}. This is not fully implemented.
Channel: List of channel files, refer to db_template('Channel').
iChannel: Index of the currently selected channel in Channel list.
Data: List of all data files, either raw or imported recordings/trials. Refer to db_template('Data') and the DataType field.
HeadModel: List of all head model files, refer to db_template('HeadModel') and db_template('HeadModelType').
iHeadModel: Index of the currently selected head model in HeadModel list.
Result: List of all source files, including links, kernels and full sources, refer to db_template('Results'). The filename of links start with "link" and actually contain two files: the kernel and the data file.
Stat: List of all statistics files, refer to db_template('Stat') and the Type field.
Image: List of all image or video files, refer to db_template('Image').
NoiseCov: Noise and data covariance files. The first file in the list is always the noise covariance, and the second file in the list is always the data covariance (if available). Refer to db_template('NoiseCov').
Dipoles: List of dipole files, refer to db_template('Dipoles').
Timefreq: List of all time-frequency files, refer to db_template('Timefreq').
Matrix: List of all matrix files, refer to db_template('Matrix').
SQLite structure: protocol.db
Starting from Brainstorm version 4.0 the protocol metadata is no longer available in memory as Matlab structures. Instead, it is saved in a SQLite relational database. This means that when the protocol is first created, an empty SQLite database is created and is populated with the metadata files in the protocol directory. Whenever metadata needs to be retrieved, it is queried from the SQLite database. While a bit slower, this gives us the ability to support concurrency which is essential for sharing protocols across contributors and running multiple jobs on the same protocol at the same time. More information here.
SQLite implements most of the SQL standard syntax, and stores all database information in a single file, the protocol.db file. Since we no longer need the metadata stored in Matlab structures, the protocol.mat is no longer used. For debugging purposes, you can explore the protocol.db file using the application DB Browser for SQLite. Concurrency is handled with regards to SQL access (i.e. if a Brainstorm instance is currently inserting metadata, another instance will not be able to access the SQLite database file until the modification is complete).
To avoid having long wait times, both due to concurrency or remote databases (when SQL queries need to be sent through a network), it is important to optimize as much as possible the database calls such that (1) active connections are closed as quickly as possible and (2) queries only return information that is absolutely required. Therefore, when writing SQL queries, make sure to only include columns that are needed (avoid SELECT * when possible) and return only rows that are needed (for example, if you only need the first row, add LIMIT 1 at the end of your query).
Despite the fact that the protocol metadata is now stored in SQLite database, the global variable GlobalData.DataBase is still available in Brainstorms and its content is fairly similar to the one described above, but with a few notable differences:
GlobalData.DataBase.ProtocolSubjects and .ProtocolStudies: These variables no longer exist as this information is stored in the SQLite database instead of in memory.
GlobalData.DataBase.ProtocolInfo: Have three additional fields:
iSubject: The ID of the subject that is currently active. Only the study used to be saved, but since study metadata is no longer in memory this avoids querying the database to get the subject of this study.
iReadFiles: Hash map containing all files that have already been "read" (selected). This allows the database to highlight files that are "new" but only until they are interacted with.
Key: FunctionalFile.Id
Value: FunctionalFile.LastModified
Database: Structure describing the information required to connect to the metadata database, refer to db_template('DatabaseInfo').
Rdbms: Relational database management system of the active database (only "sqlite" supported).
Location: Location of the database. For SQLite: the path to the protocol.db file.
Name: Name of the database to use (not applicable for SQLite).
Username: Username to connect to the database (not applicable for SQLite).
Password: Password to connect to the database (not applicable for SQLite).
SQL Schema
A SQL schema defines all the tables, the columns of every table and the relation between tables in the database. Refer to the sql_generate_db() function: the name, type and NOT NULL properties of the tables columns are defined in db_template(), and additional properties like foreign keys and primary keys are hardcoded in sql_generate_db().
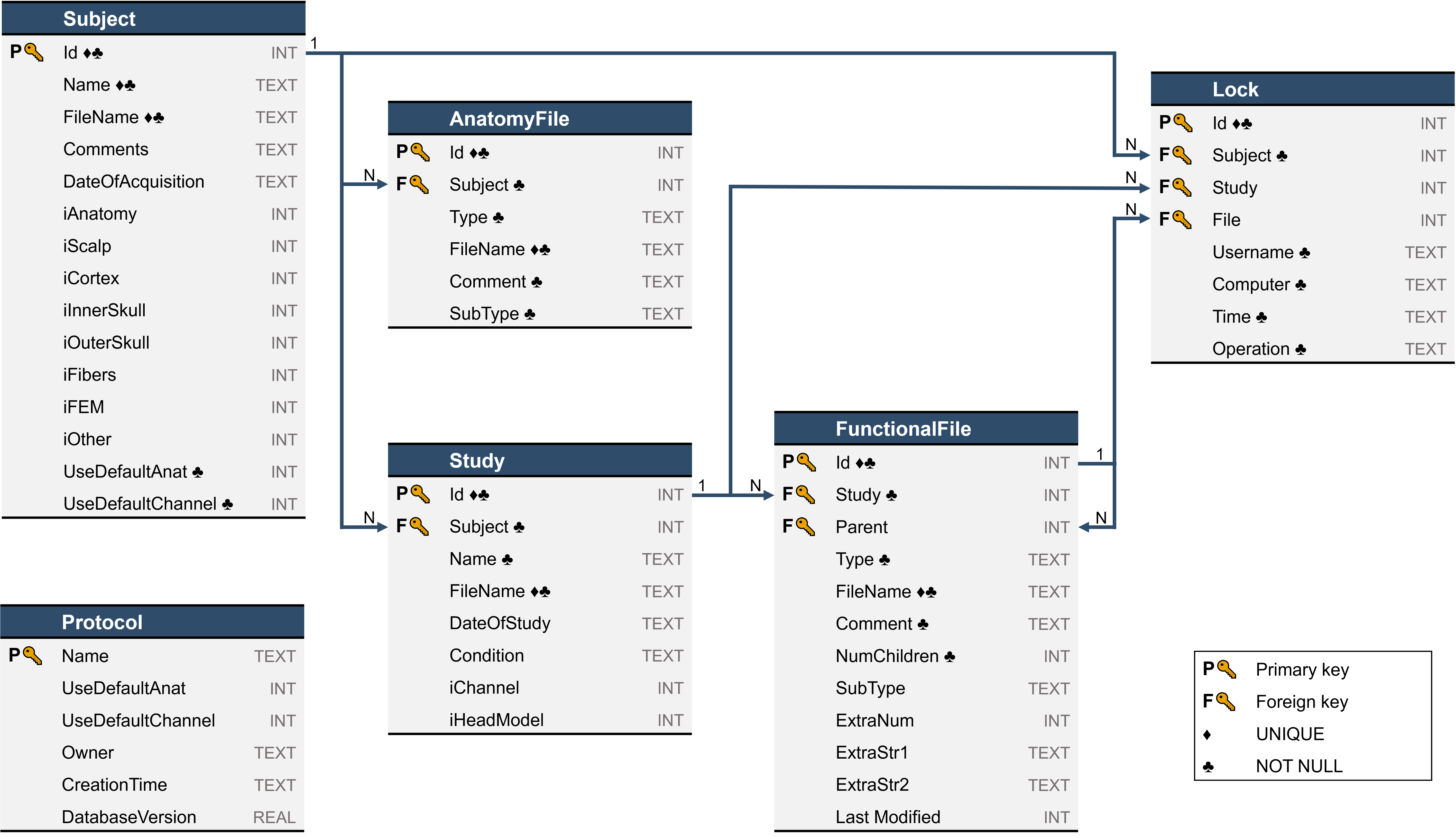
Below a description of the tables in the SQLite database.
Protocol: Table containing the metadata for the current protocol. It always has a single row!
Name: Name of the protocol.
UseDefaultAnat: Same as GlobalData.DataBase.ProtocolInfo.
UseDefaultChannel: Same as GlobalData.DataBase.ProtocolInfo.
Owner: The PC username of the person that created this protocol, refer to bst_get('UserName').
CreationTime: When this protocol was created.
DatabaseVersion: Version of this database schema. If new fields are added to the database in subsequent versions, old protocols should be detected using this field and their schema should be updated.
Subject: Table containing the metadata for all subjects. Very similar to GlobalData.DataBase.ProtocolSubjects in the previous metadata structure, with a few notable differences:
Id: Unique identifier number for each subject. This is used to link this table with other tables (foreign keys).
iAnatomy, iCortex, etc.: These indices are now unique AnatomyFile IDs rather than the position in the list of all anatomy/surface files.
Anatomy, Surface: These lists no longer exist and are represented as entries in the AnatomyFile table instead.
AnatomyFile: Table containing the metadata for all anatomy files (MRI, surface, fibers, etc), refer to db_template('AnatomyFile').
Id: Unique identifier number for each anatomy file. This is used to link this table with other tables (foreign keys).
Subject: ID of the subject this file belongs to (foreign key).
Type: Type of anatomy file ("volume", "surface"). Used to split back into Anatomy and Surface lists if needed.
FileName: Relative path to the anatomy file.
Comment: Name of the entry in the database.
SubType: Additional field to identify the subtypes of volumes and surfaces.
Study: Table containing the metadata for all studies. Very similar to ProtocolStudies in the previous metadata structure, with a few notable differences:
Id: Unique identifier number for each study. This is used to link this table with other tables (foreign keys).
Subject: ID of the subject this study belongs to (foreign key).
iChannel and iHeadModel: These indices are now unique FunctionalFile IDs rather than the position in the list of all Channel/HeadModel files.
Channel , Data , etc: These lists no longer exist and are represented as entries in the FunctionalFile table instead.
FunctionalFile: Table containing the metadata for all functional files (data, sources, etc), refer to db_template('FunctionalFile').
Id: Unique identifier number for each functional file. This is used to link this table with other tables (foreign keys).
Study: ID of the study this file belongs to (foreign key).
Parent: The ID of the parent functional file this file belongs to, or NULL if it's directly under the study.
Type: Type of functional file (data, matrix, result, etc).
FileName: Path to the functional file.
Comment: Name of the file in the database.
NumChildren: Number of child files (i.e. the number of files where ParentFile = this ID). This is used to display the number of files in a list and avoids COUNT queries that might slow down displaying the database.
SubType: Some file types have sub types, they are defined here. E.g.: data -> raw vs recordings.
ExtraNum / ExtraStr1 / ExtraStr2: Additional numerical or string columns used by specific file types. These are defined in db_get().getFuncFileStruct().
LastModified: Unix time timestamp of the last time this file was modified.
Lock: Table containing all active locks on protocol entities, refer to db_template('Lock').
Id: Unique identifier number for this lock.
Subject: ID of the subject this entity belongs to (foreign key).
Study: ID of the study this entity belongs to (foreign key), or NULL if the lock is on the whole subject.
File: ID of the locked functional file (foreign key), or NULL if the lock is on the whole subject or study.
Username: PC username of the person that acquired this lock, refer to bst_get('UserName').
Computer: PC name of the person that acquired this lock, refer to bst_get('ComputerName').
Time: UTC Date & time when this lock was acquired.
Operation: Description of the operation that acquired this lock, typically the process name.
SQLite API in Brainstorm
This API consists on functions that wrap the functionality of SQL queries to read and write the protocol metadata SQLite database. These functions are divided in two types according to their abstraction level.
High level
The most important functions to read and write the SQLite database are:
db_get(): Get (read) data from the DB. It presents useful cases. It is similar to bst_get().
db_set(): Set (write) data from the DB. It presents useful cases. It is similar to bst_set().
A simple example of using db_get and db_set
% Get all info for default subject with Id = 1
sSubject = db_get('Subject', 1);
% Update the name of this subject.
sSubject.Name = 'J.Doe';;
db_set('Subject', sSubject);
Low level
The db_get and db_set functions rely in lower level functions that interact with the database:
sql_connect(): To get an active connection to the database. It also sets the PRAGMAs that are needed for each connection. Optional argument of type db_template('DatabaseInfo') to define how to connect to the database, otherwise it looks into the active ProtocolInfo.
sql_close()(sqlConnection): Close an active connection to the database.
sql_create()(sqlConnection, tables): Generate and executes SQL CREATE queries for every table as input. Tables input structure defined in db_template('sqltable').
sql_query(): This is versatile function that:
- Execute direct queries created by the user.
Execute "SELECT", "EXIST", "INSERT", "UPDATE", "DELETE" and "COUNT" SQL queries to the SQLite database. For the proper syntax, check the documentation of sql_query().
- Can handle the opening and closing of the database if it's necessary.
Here are some specific SQL query examples:
1. Direct query
This allows you to write manual SQL queries and execute them directly. The drawback is that it returns a java.sql.ResultSet object so you need to be familiar with this specific syntax, and you need to handle the SQL connection yourself. Example:
sqlConnection = sql_connect();
resultSet = sql_query(sqlConnection, 'SELECT * FROM Subject');
while resultSet.next()
disp(resultSet.getString('Name'));
end
resultSet.close();
sql_close(sqlConnection);2. SELECT query
This lets you read data from the database. It generates a SELECT query, executes it for you, and returns a db_template() structure of the proper table. Example:
% Get all info of subjects that use the default anatomy.
sSubject = sql_query('SELECT', 'Subject', '*', struct('UseDefaultAnat', 1));
% Get the name of the first subject.
sSubject = sql_query('SELECT', 'Subject', 'Name', [], 'LIMIT 1');
% Get the name and ID of all subjects.
sSubject = sql_query('SELECT', 'Subject', {'Id', 'Name'});3. UPDATE query
This lets you modify rows in the database. It generates an UPDATE query, executes it for you, and returns the number of rows modified. Example:
% Makes subject #1 use the default anatomy
numRows = sql_query('UPDATE', 'Subject', struct('UseDefaultAnat', 1), struct('Id', 1));
% Makes all subjects use the default anatomy
numRows = sql_query('UPDATE', 'Subject', struct('UseDefaultAnat', 1));4. INSERT query
This lets you insert rows into the database. It generates an INSERT query, executes it for you, and returns the generated ID of the inserted rows. It is strongly recommended to use db_template() to define the values to insert. Example:
sSubject = db_template('Subject');
sSubject.Name = 'MyNewSubject';
SubjectId = sql_query('INSERT', 'Subject', sSubject);5. DELETE query
This lets you delete rows from the database. It generates a DELETE query, executes it for you, and returns the number of rows deleted. Be careful, this cannot be undone! Example:
% Delete Subject #1
numRows = sql_query('DELETE', 'Subject', struct('Id', 1));
% Delete all subjects
numRows = sql_query('DELETE', 'Subject');