[TUTORIAL UNDER CONSTRUCTION]
FEM tutorial: MEG/EEG Median nerve stimulation
Authors: Takfarinas Medani, Juan Garcia-Prieto, Wayne Mead, Francois Tadel
This tutorial introduces the FEM modeling in the Brainstorm environment, using DTI tensors and third-party programs BrainSuite, SimNIBS and CAT12.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Contents
Dataset description
Experiment
The experiment consists of an unilateral median nerve stimulation conducted in a MEG laboratory with an Elekta Triux (Megin, Finland) scanner.
- The stimulation signal was a square-wave pulse with 2Hz frequency and duration of 0.2ms.
- An ISI (inter-stimulus interval) of 500ms with a variation of ±20ms in order to be able to average out time-locked noise to the stimulation, while remaining unnoticeable by the subject.
- The stimulation was performed on the left hand/wrist, for two minutes, with a Digitimer DS7A stimulator. An electrode was placed on the hand/wrist while current value was tuned to match the motor threshold of the subject on the stimulated hand, with a result of 10~12mA approximately.
- Recordings were performed with a 1kHz sampling rate. Continuous HPI was disabled during these recordings. High-pass filters were set to DC for MEG channels and 0.03Hz for EEG channels.
All MEG files underwent a MaxFilter (version 2.3.13) tsss post-processing.
MRI imaging scanning (Philips Medical Systems):
- T1w image without contrast: 3T field strength, flip angle 8°, TR 7.9s, dimensions 512 × 512 × 200, and voxel dimensions of 0.469 × 0.469 × 0.939 millimeters
- T2w spin-echo image: 3T field strength, 78.57% phase FOV, 90° flip angle, SAR 0.327, voxel dimensions 560 × 560 × 55, dimensions of 0.429 × 0.429 × 3 millimeters
- DWI sequences: voxel dimensions 512 x 512 x 200 and dimensions of 0.469 × 0.469 × 0.939 millimeters, diffusion-sensitizing gradients in 32 non-collinear directions
Files
The dataset we distribute with this tutorial follows the Brain Imaging Data Structure (BIDS) standard for neuroimaging data organization. The files that will be imported in this tutorial are the following:
sample_fem/sub-fem01/: Raw data for subject fem01
ses-meg/: Simultaneous recordings of MEG and EEG.
sub-fem01_ses-meg_task-mediannerve_run-01_proc-tsss_meg.fif
ses-mri/: Imaging exams.
anat/sub-fem01_ses-mri_T1w.nii.gz: T1-weighted MRI
anat/sub-fem01_ses-mri_T2w.nii.gz: T2-weighted MRI
dwi/sub-fem01_ses-mri_dwi.*: Diffusion-Weighted Imaging (DWI)
License
Creative Commons CC0 1.0 Universal. This dataset is distributed in the public domain.
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Additional programs: You need to install the following programs to follow this tutorial:
SimNIBS: Description | Download
BrainSuite: Description | Download
MATLAB Runtime: Follow the instructions on the BrainSuite download page
SPM12: Install as Brainstorm plugins
CAT12: Install as Brainstorm plugins
Iso2mesh: Install as Brainstorm plugins
Download the dataset:
Go to the Download page and get the file: sample_fem.zip
- Unzip it outside of any of the Brainstorm folders (program folder or database folder)
Brainstorm:
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialFem" and select:
"No, use individual anatomy"
"No, use one channel file per condition".
Import the anatomy
T1 MRI
- Switch to the "anatomical data" view, the left button in the toolbar above the database explorer.
Right-click on the TutorialFem folder > New subject > Subject01
- Keep the default options you set for the protocol.
Right-click on the subject node > Import MRI:
Set the file format: All MRI files (subject space)
Select the T1 file: sub-fem01/ses-mri/anat/sub-fem01_ses-mri_T1w.nii.gz
Click on the link "Click here to compute MNI normalization": option "maff8".
This estimates an affine transformation to the MNI space and sets default positions for the anatomical fiducials. Because we will use the digitized head shape of the subject to refine the MRI/MEG registration, we don't need the position of the fiducials to be very accurate.
T2 MRI
Right-click on the subject node > Import MRI:
Set the file format: All MRI files (subject space)
Select the T2 file: sub-fem01/ses-mri/anat/sub-fem01_ses-mri_T2w.nii.gz
- Brainstorm asks about registering the new T2 with the reference T1 image:
Select the option: SPM
Reslice the volume: Yes
This process will take few minutes, just be patient.
Diffusion imaging
The FEM has the ability to incorporate anisotropic conductivity from MRI diffusion imaging, which is particularly interesting for the modeling of the white matter. Brainstorm can load the Diffusion-Weighted Images (DWI), and compute the tensors (DTI) using the BrainSuite Diffusion Pipeline (BDP). This requires BrainSuite to be installed on your computer, with the bdp program available in the system path.
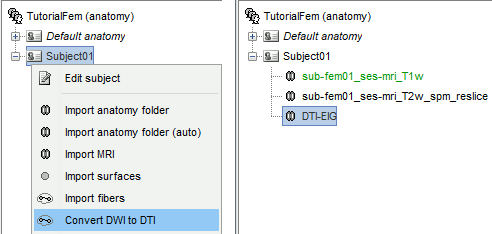
Right-click on Subject01 > Convert DWI to DTI
Select the DWI file: sub-fem01/ses-mri/dwi/sub-fem01_ses-mri_dwi.nii.gz
- The associated files *dwi.bvec and *dwi.bval must be in the same folder.
The process can take up to 30min. At the end, a new file DTI-EIG appears in the database. This file contains 12 volumes, ie. 12 values for each voxel. From 1 to 9: components of the three eigenvectors; from 10 to 12: the values of their norm to the eigenvalue.
FEM head model
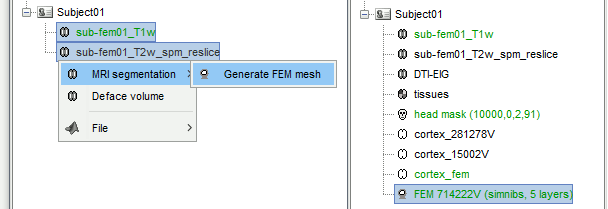
The FEM approach requires a segmentation of the head volume in different tissues, represented as hexahedral or tetrahedral 3D meshes. The methods available within Brainstorm are listed in the tutorial FEM mesh generation. This tutorial illustrates only the use of SimNIBS.
FEM mesh with SimNIBS
Running SimNIBS:
- Select both the T1 and the T2 MRIs, using the mouse and the SHIFT or CTRL keys.
Right-click on the selected files > MRI segmentation > Generate FEM mesh > SimNIBS. Keep the default options.
- This process can take 2-5 hours.
- The output of SimNIBS is saved in the temporary folder $HOME/.brainstorm/tmp/simnibs, and then imported into the Brainstorm database.
At the end of this computation, new files are available in the database:
tissues: Segmentation of the head volume in 6 different tissues
head mask: Outer surface of the scalp
cortex_280000V: Pial surface generated by CAT12 within the SimNIBS process
cortex_15002V: Low-resolution version of the pial surface
cortex_fem: Outer surface mesh of the gray matter, extracted from the FEM mes
FEM 70000V: Tetrahedral FEM mesh with 5 layers (gm, wm, CSF, skull, scalp).
At the end of the process, make sure that the file "cortex_15002V" is selected (downsampled pial surface, which will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.
Remesh with Iso2mesh
In some cases, the FEM mesh generated with SimNibs causes issues with the DUNEuro FEM solver, due to the air cavities that are not tesselated. In order to avoid these possible issues in the next steps, we will correct the mesh using the Iso2mesh functions integrated within Brainstorm.
Richt click on the FEM mesh > Extract surfaces.
- Hold the CTRL key to select all the extracted surfaces (white, gray, csf, skull, scalp).
Right-click > Generate FEM mesh > Iso2mesh-2021 > MergeMesh > Max volume = 0.001, Percentage kept = 100. Note that the higher the resolution leads to higher accuracy but also higher memory requirements and longer computation times.
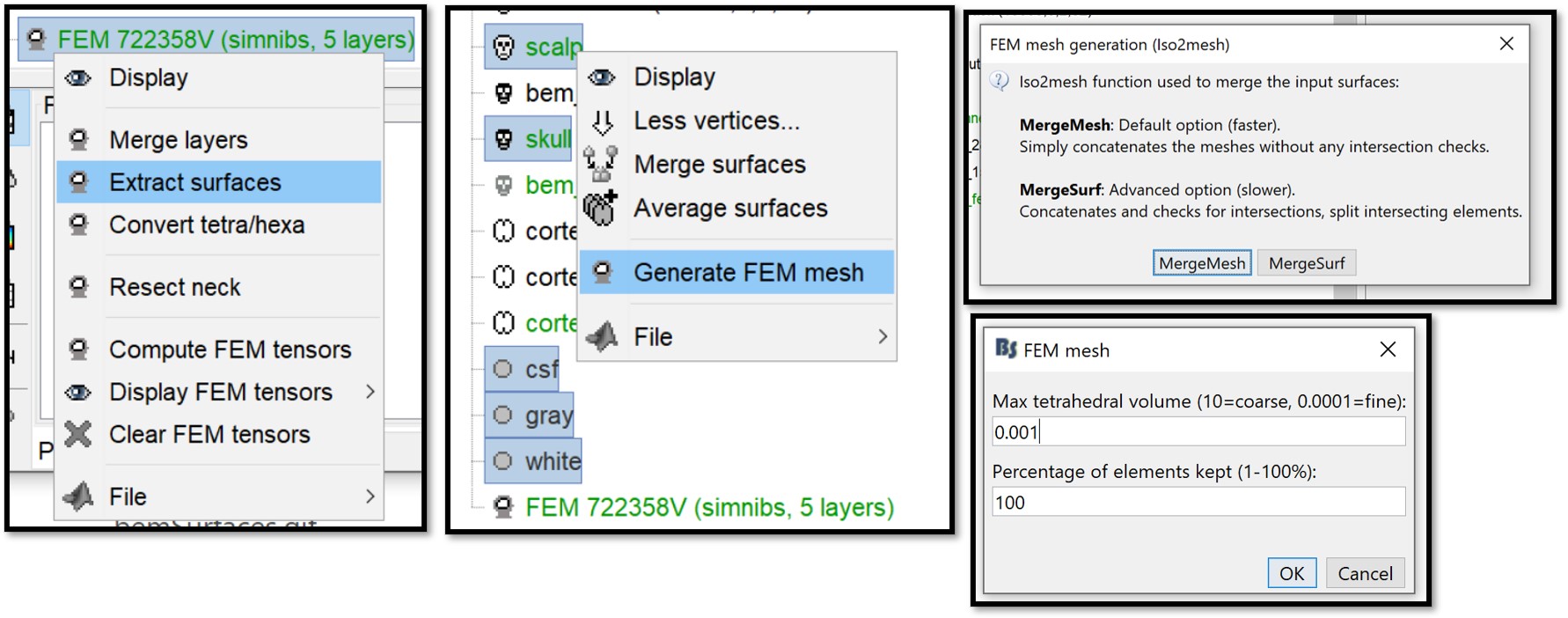
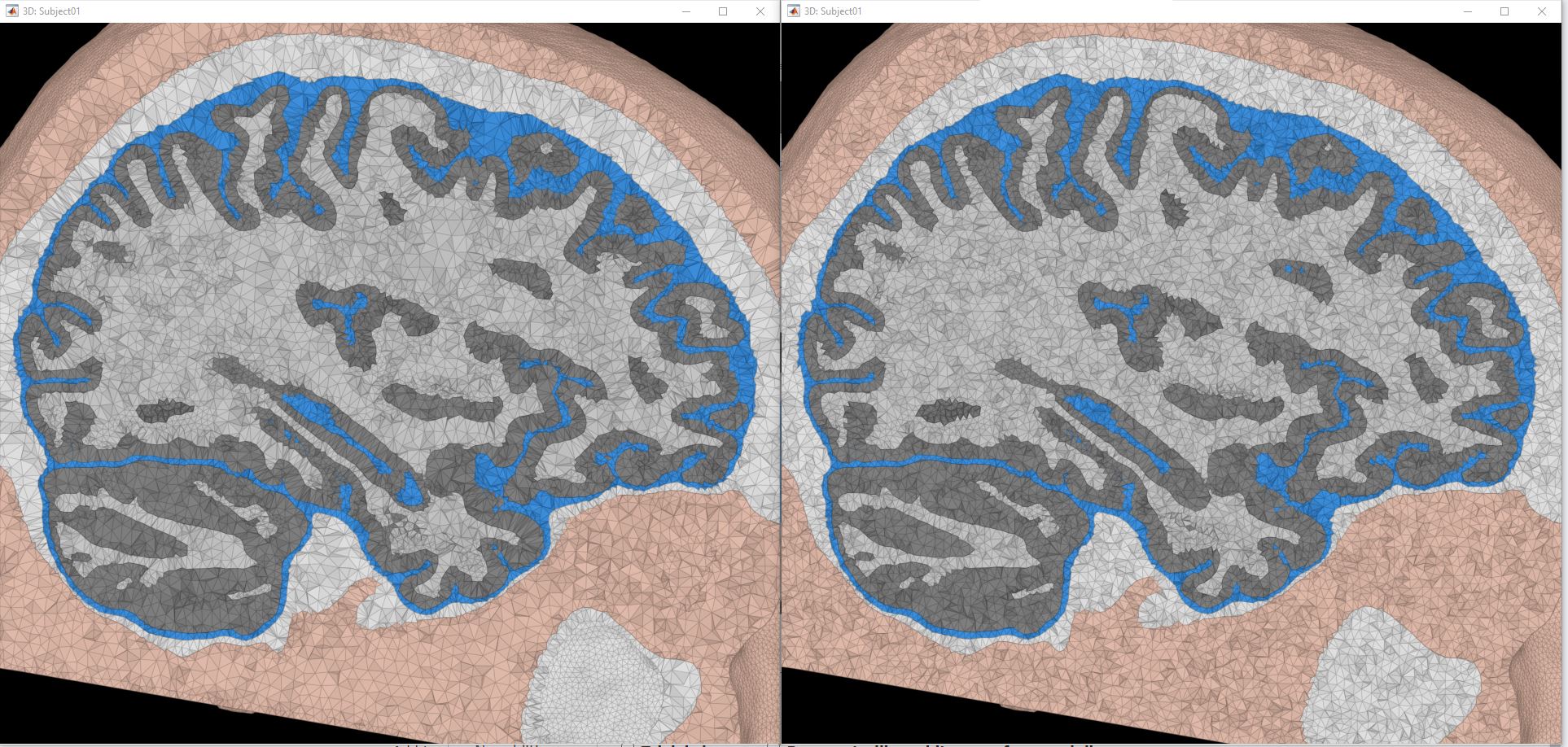
The following figure shows the two model, left is the initial model obtained with SimNibs, right is the second model obtained from the Iso2Mesh remesh: the air cavity in the bottom-right corner is now fully tesselated. Note that you can also use this process to generate FEM models with higher mesh densities.
FEM tensors
We can now incorporate the diffusion information into the FEM model, and compute anisottropic conductivity tensors for the tetrahedral elements of the white matter (we consider the other tissues to have isotropic conductivities).
Right-click on the FEM mesh > Compute FEM tensors. Select the options as illustrated below, and refer to the tutorial FEM tensors for more information.
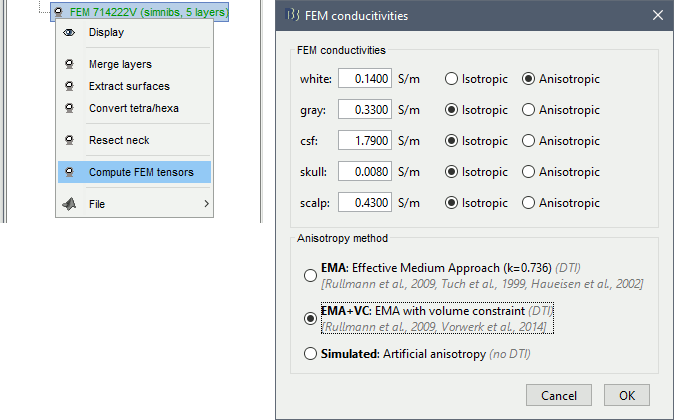
- This process can take up to 5min, depending on the resolution of the FEM mesh.
Visualization
Brainstorms include the possibilities to display the FEM head models and the tensors, users can also overlay the display with the MRI as well as with the different surfaces.
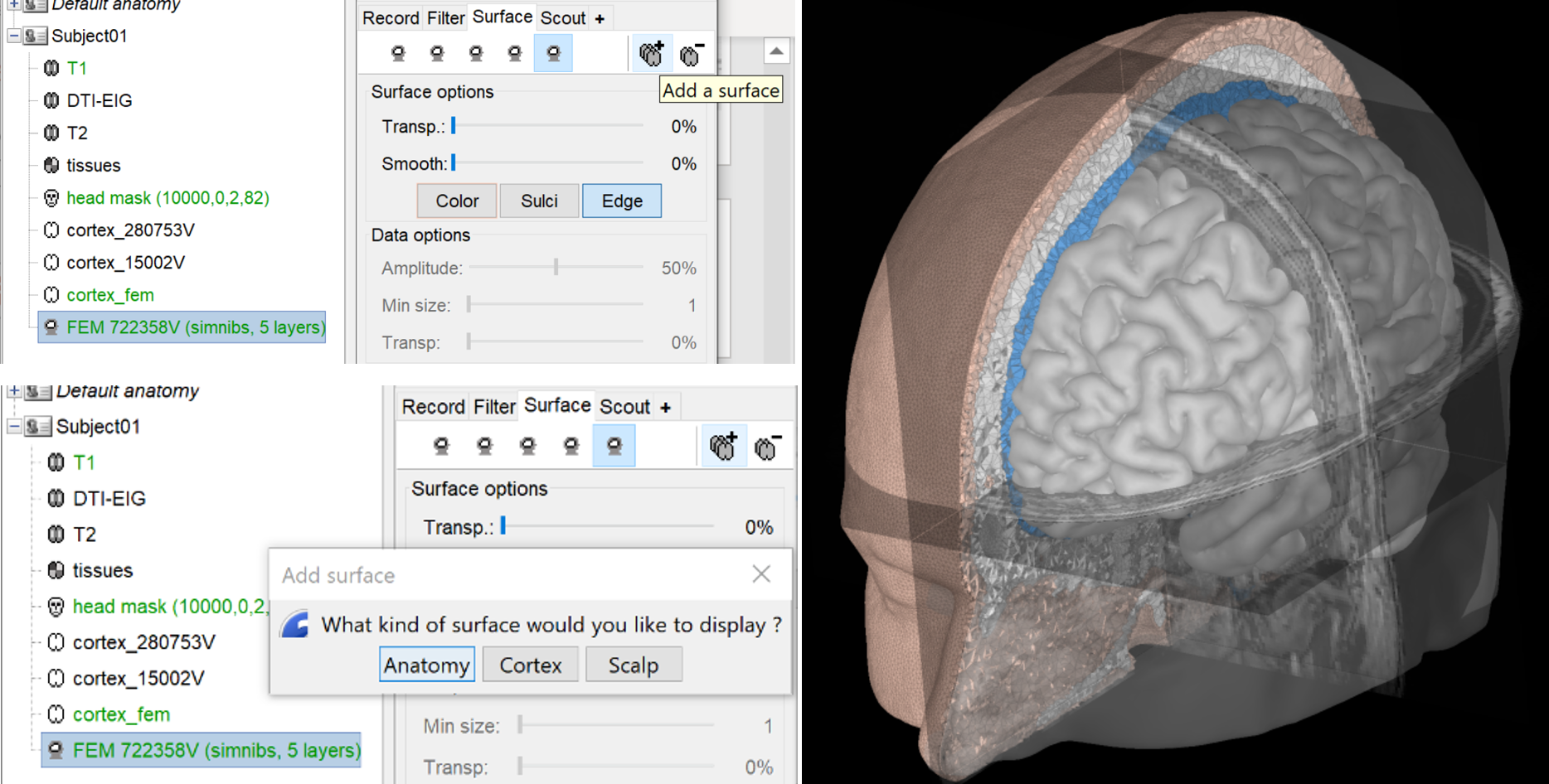
Right-click on the FEM mesh > Display FEM tensors: The FEM tensors can be displayed on the mesh or MRI, as arrows on the main eigenvector or as ellipsoids on each FEM element.
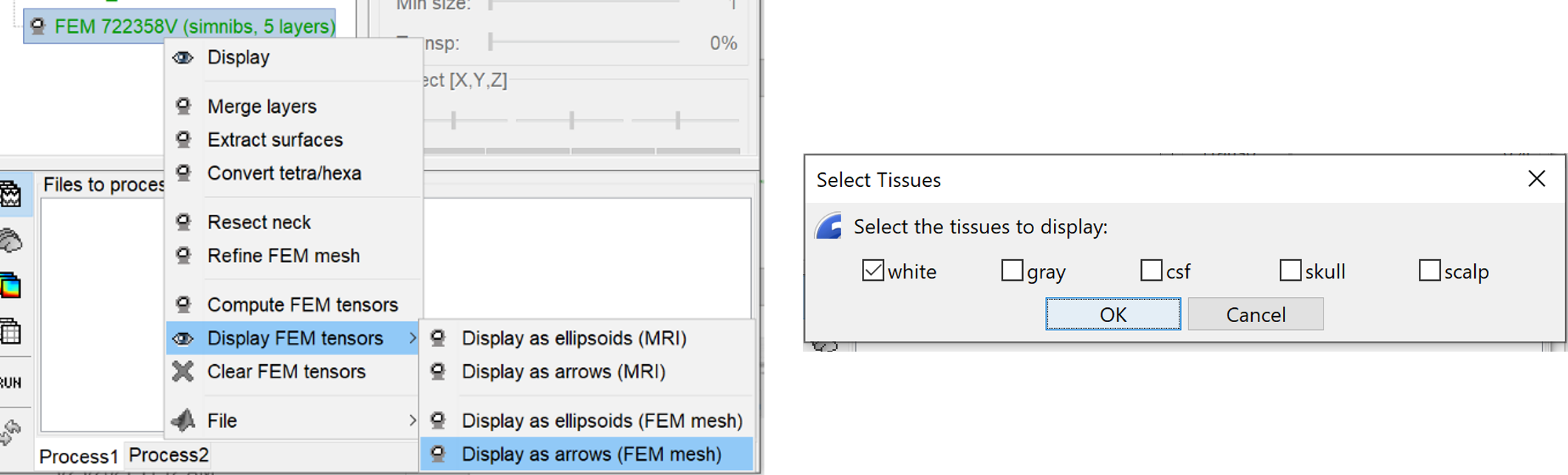
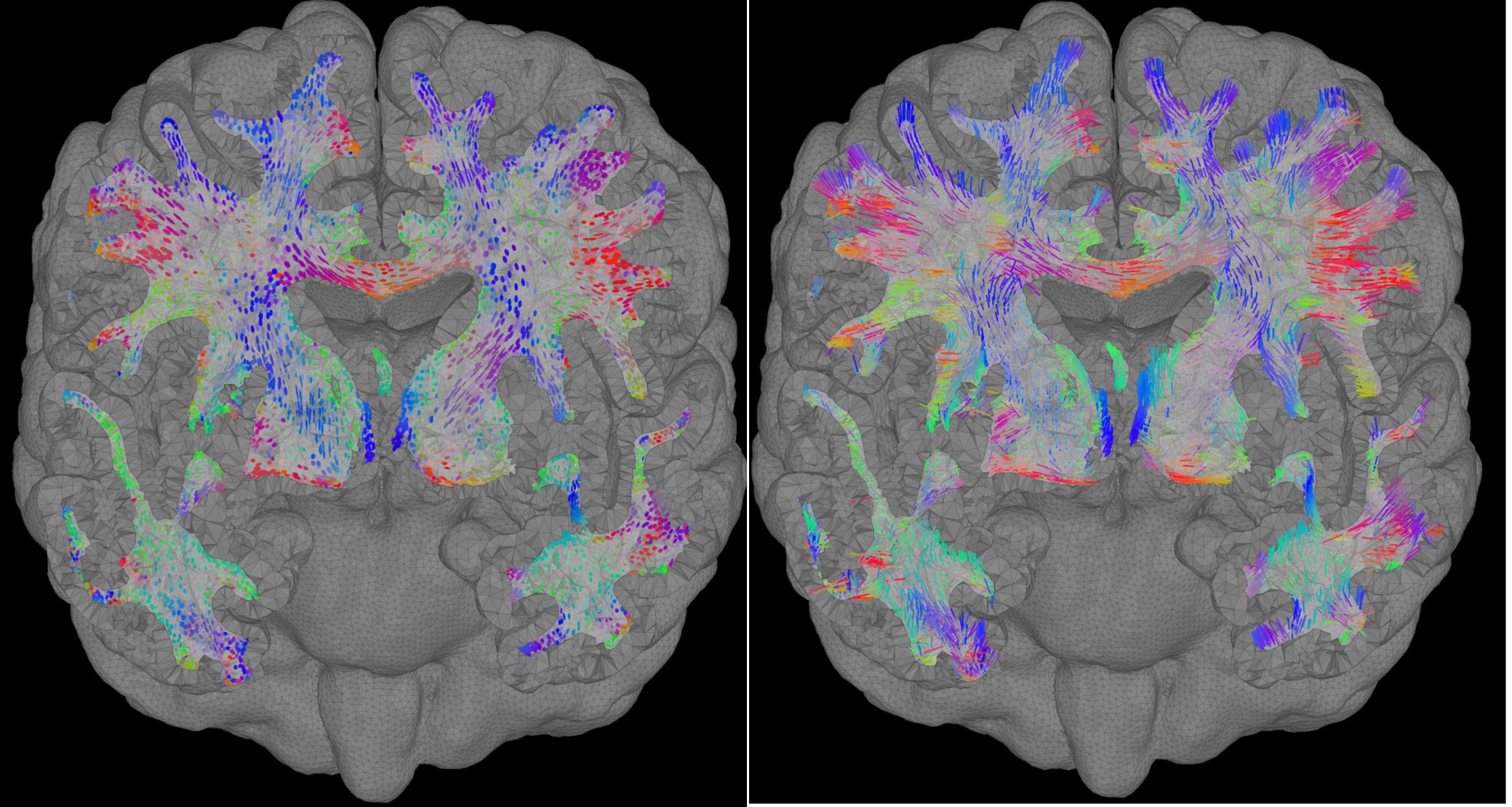
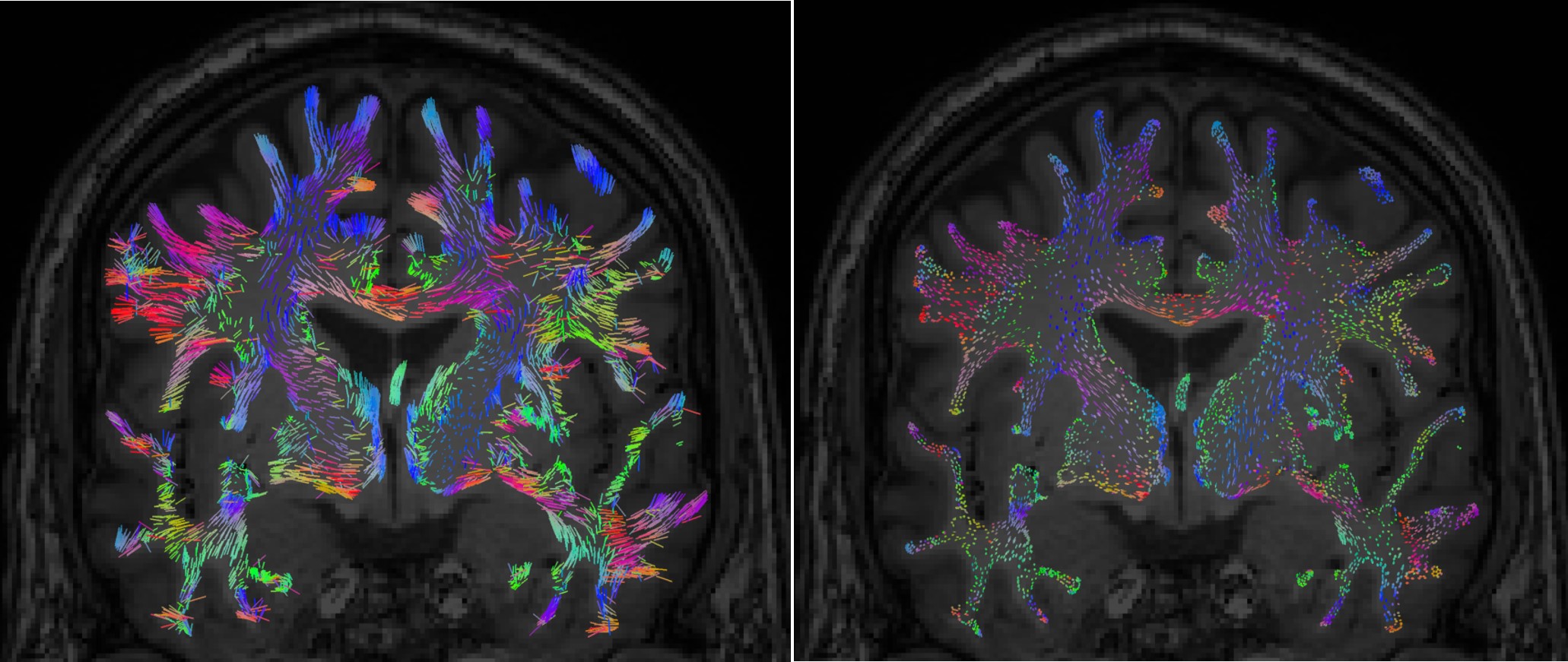
To configure the display: right-click on the figure > FEM tensors menu. The keyboard shortcuts for changing the size of the displayed tensors are the Up and Down arrows keys. You can also switch the display mode by using the shortcut "Shift + Space".
On the hard drive
The DTI-EIG file as the same structure as any MRI file, with 12 volumes stacked along the 4th dimension. From 1 to 9: components of the three eigenvectors; from 10 to 12: the values of their norm to the eigenvalue.
The FEM mesh contains the following fields:
Comment: String displayed in the database explorer to represent the file.
Vertices: [Nvertices x 3], coordinates (x,y,z) of all the nodes of the tetrahedral mesh, in SCS coordinates.
Elements: [Nelements x 4], list of indices corresponding to the rows of the Vertices matrix, each set of 4 connected vertices indentifying a tetrahedron of the mesh.
Tissue: [Nelements x 1], for each Element, index of the corresponding tissue (e.g. wm=1, gm=2, csf=3, skull=4, scalp=5)
TissueLabels: Labels of the tissues, e.g. {'white' 'gray' 'csf' 'skull' 'scalp'}
Tensors: [Nelements x 12] if tensors are computed, otherwise empty. The 12 values are the eigenvalues and eigenvectors interpolated on each element of the mesh.
History: List of operations performed on this file (menu File > View file history).
BEM head model
For the purpose of comparison between the FEM and the BEM, we will generate also the BEM surfaces for this subject and we will follow the same step as explained in the BEM tutorial.
Right-click on Subject01 > MRI segmentation > Generate BEM surfaces > Brainstorm.
Keep the default options: 1922 vertices for all the layers, skull thickness = 4m.- Three new files appear in the database explorer: bem_head_1922V, bem_outerskull_1922V, and bem_innerskull_1922V. These surfaces will be used for the BEM forward computation.
Now we have two head geometry available for this subject, BEM and FEM
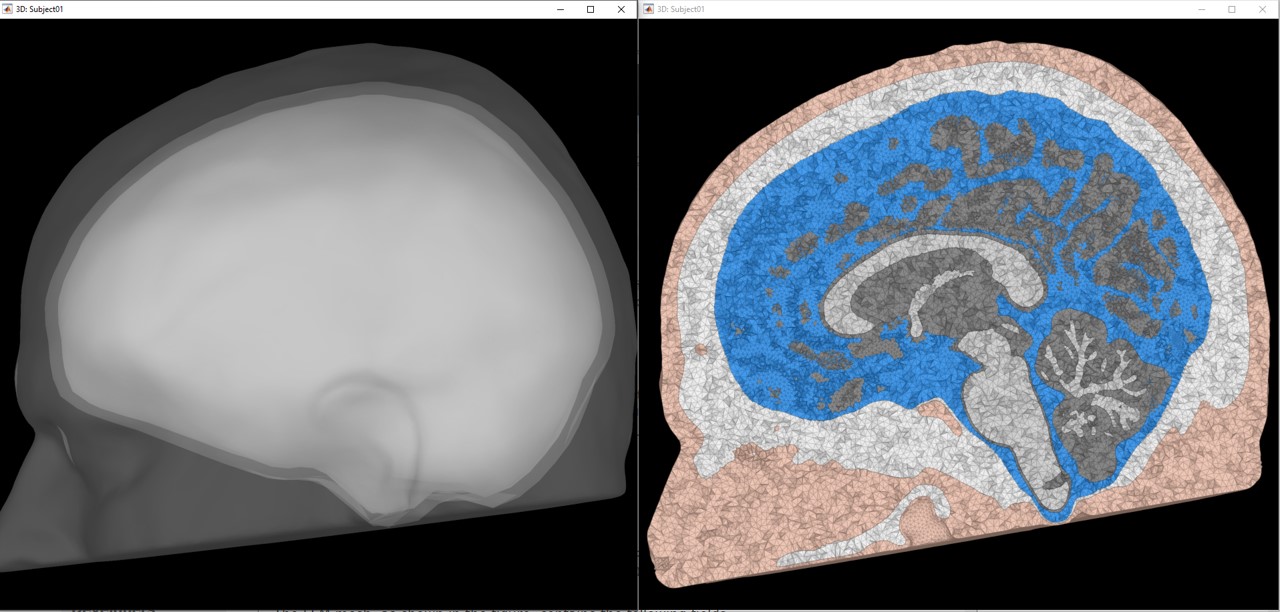
Access the recordings
Link the recordings
- Switch to the "functional data" view, the middle button in the toolbar above the database explorer.
The folder "eeg_positions" which contains contains default 10-10 EEG electrodes positions, was generated by the SimNIBS process. We will not use it in this tutorial: delete this folder.
Right-click on the subject folder > Review raw file:
- Select file type: MEG/EEG: Electa-Neuromag(*.fif)
Select file: sub-fem01/ses-meg/meg/sub-fem01_ses-meg_task-mediannerve_run-01_proc-tsss_meg.fif
FIF event file: Select Event Channel, this will read the events markers
Refine registration? YES
A figure is opened to show the registration MRI/MEG.
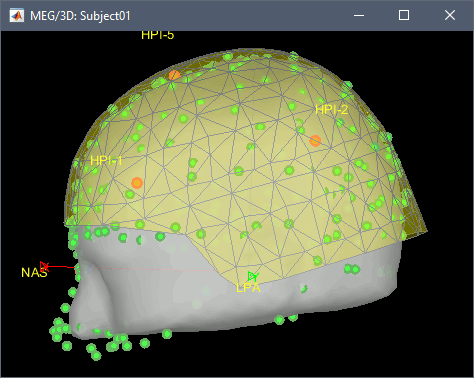
- The new file "Link to raw file" lets you access directly the contents of the MEG/EEG recordings. The "Neuromag channels (374)" contains the name of the channels and the position of the corresponding sensors (MEG/EEG).
Channel file
- The recordings contain signals coming from different types of electrodes:
- 306 MEG channels (102 magnetometers and 204 gradiometers )
- 64 EEG channels
- 2 EOG channels: EO1 and EO2
- 1 Stim channel that contain the stimulation signals
Right-click on the channel file > MRI registration > Edit... (EEG)
- The white points are the electrodes, the green points are the additional digitized head points. To display the label of the electrodes, click on the [LABEL] button in the toolbar. To see what the other buttons in the toolbar are doing and how to use them, leave your mouse over them for a few seconds and read the description.
- The automatic registration based on the head points is already good, therefore we won't apply any additional rotation+translation to the MEG+EEG sensors.
Click on the button "Project electrodes on scalp surface", it will help for the source modeling later. The green points (digitized) stay in place, the white points (electrodes) are now projected on the skin of the subject.
- If you feel like you didn't do this correctly, close the figure and cancel the modifications, then try again.
Click on [OK] when done. Answer YES to save the modifications.
Pre-processing
- Drag and drop the "Link to raw file" into the Process1 list.
Select the process "Frequency > Power spectrum density", configure it as in the figure
After the computation, double-click on the new PSD file to display it.
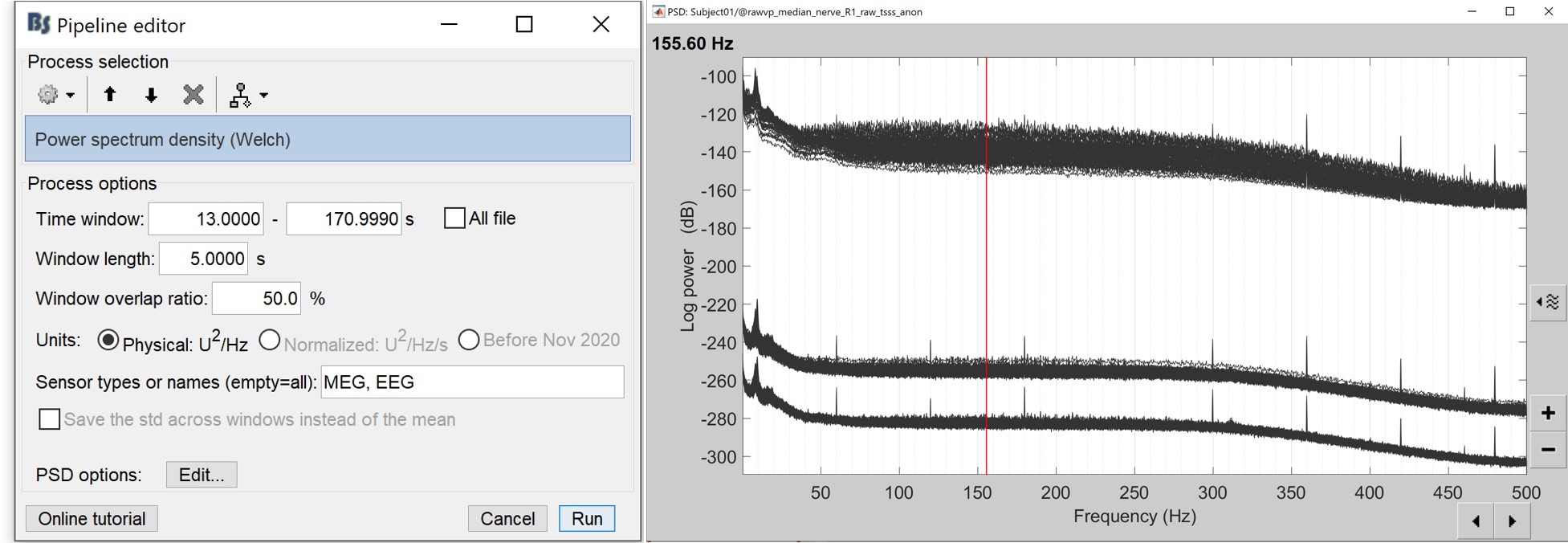
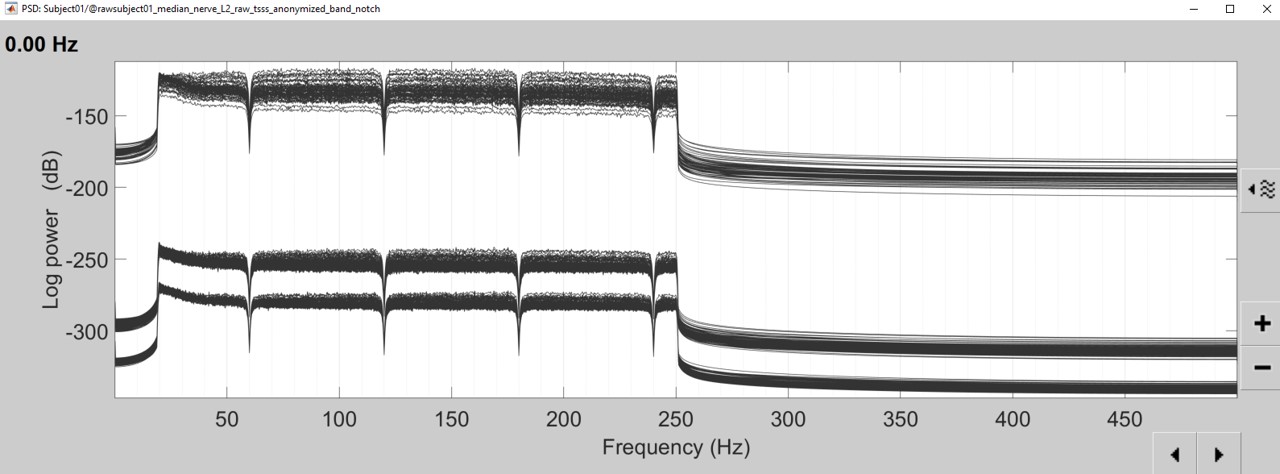
- The lines on the top represent the EEG electrodes, the lines at the bottom the MEG sensors(MAG and GRAD). If you want to get clearer plots, you can calculate separately the spectrum for the two types of sensors separately, by running twice the process "Power spectrum density", once with sensor types = "MEG" and once with "EEG", instead of running in on both at the same time as we did.
- Observations (below 250Hz):
- Peak around 10Hz: Alpha waves from the subject's brain
- Peaks at 60Hz, 120Hz, 180Hz, 240Hz on EEG + MEG: Power lines (60Hz+harmonics)
- Most of the signal looks clean and no bad channel is observed
- If we review quickly the EEG and EOG signals, we notice that they are quite clean and no artifacts are observed. The experiment is well designed and the subject was not moving the eyes (blinks and slow movements), maybe because there was a fixation cross for this experiment. However, we will apply at least a high-pass filter to make the signals easier to process (we are not interested in very low frequencies in this experiment).
Frequency filters
- In Process1, select the "Link to raw file".
Select process Pre-process > Band-pass filter: Frequency=[20, 250]Hz, Sensors=MEG,EEG
Add processs Pre-process > Notch filter: Frequencies=[60 120 180]Hz, Sensors=MEG,EEG
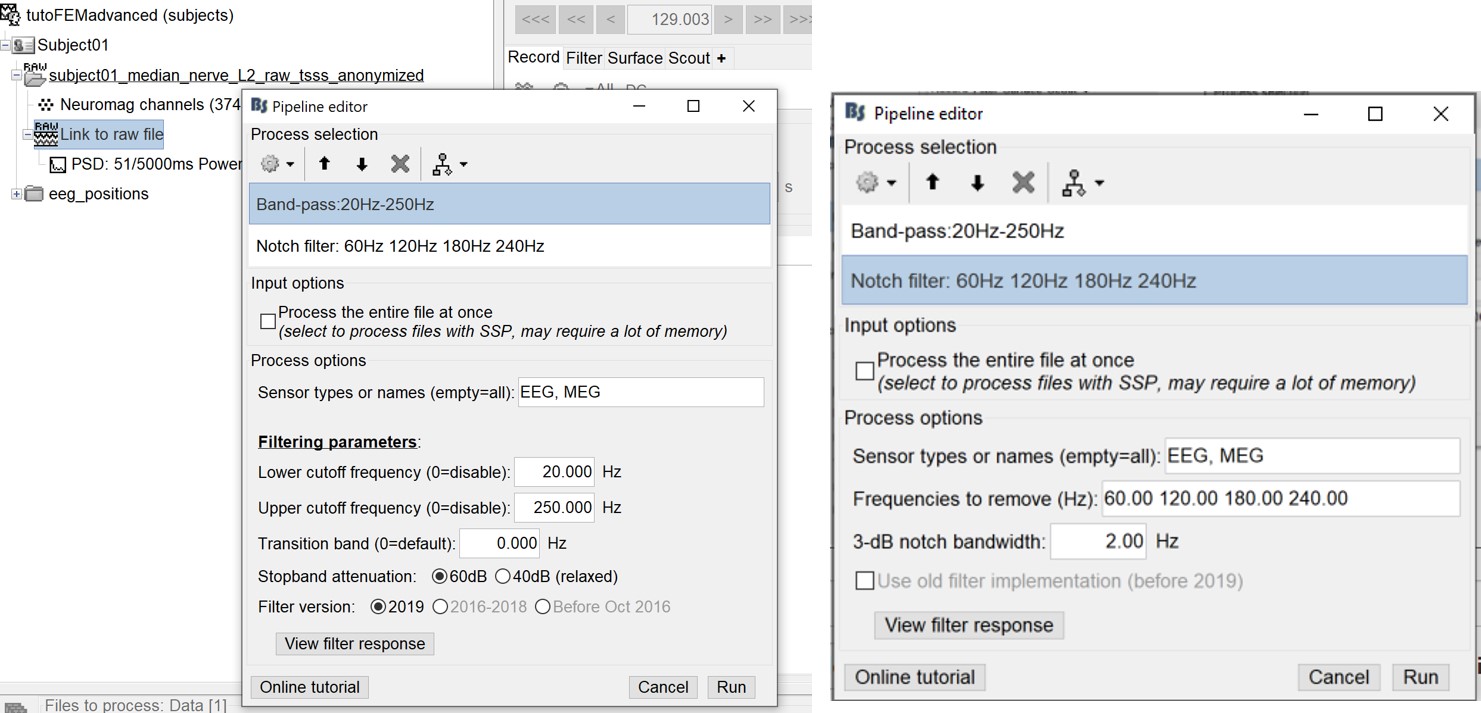
- In Process1, select the filtered file "Raw | Band | Notch".
Run the process "Frequency > Power spectrum density", with the same options as before. And compare with the previous figure. You may find that the power line frequency and its harmonics are removed and the data is filtered between 20 and 250Hz.
- To keep the interface clean and easy to follow: keep only the last continuous file (Raw|Band|Notch). Delete the link to the original file (which will not delete the original .fif file), and delete the "Raw|Band" file.
EEG: Average reference
Right-click on the filtered file "Raw|band|notch" > EEG > Display time series.
In the Record tab, menu Artifacts > Re-reference EEG > "AVERAGE".
At the end, the window "Select active projectors" is open to show the new re-referencing projector. Just close this window. To get it back, use the menu Artifacts > Select active projectors.
Epoching
In this experiment, the electric stimulation is sent with a frequency of 2Hz, meaning that the inter-stimulus interval is 500ms. We are going to import epochs of 300ms around the stimulation events (-100 to 200ms).
Right-click on the filtered file "Raw|band|notch" > Import in database:
Check "Use events" and select "2", this is the name that brainstorm assigns for the stimulus onset, it may have different name on your computer. Set epoch time: [-100, 200] ms and Apply SSP/ICA in order to apply the average reference.
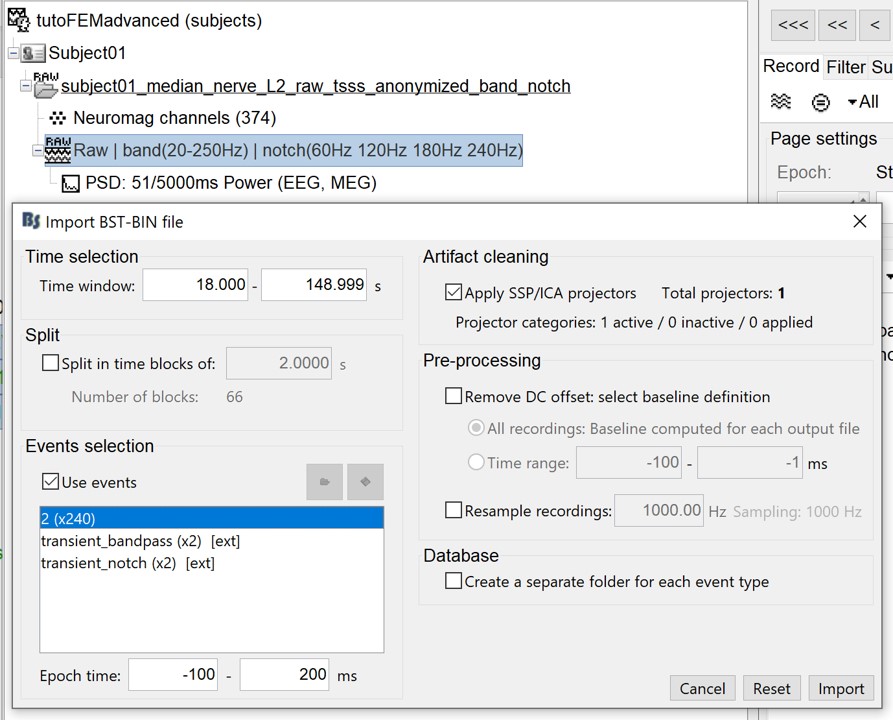
- At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,200]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment (240), we can just ignore it. The total number of epochs is then 239.
Averaging
- Drag and drop all the Trigger01 trials to the Process1 tab.
Run the process Average > Average files: By trial group (folder average)
Review the average for the MEG and EEG as a topography plot: Right-click on the averaged signal > EEG > 2D disc, repeat for MEG.
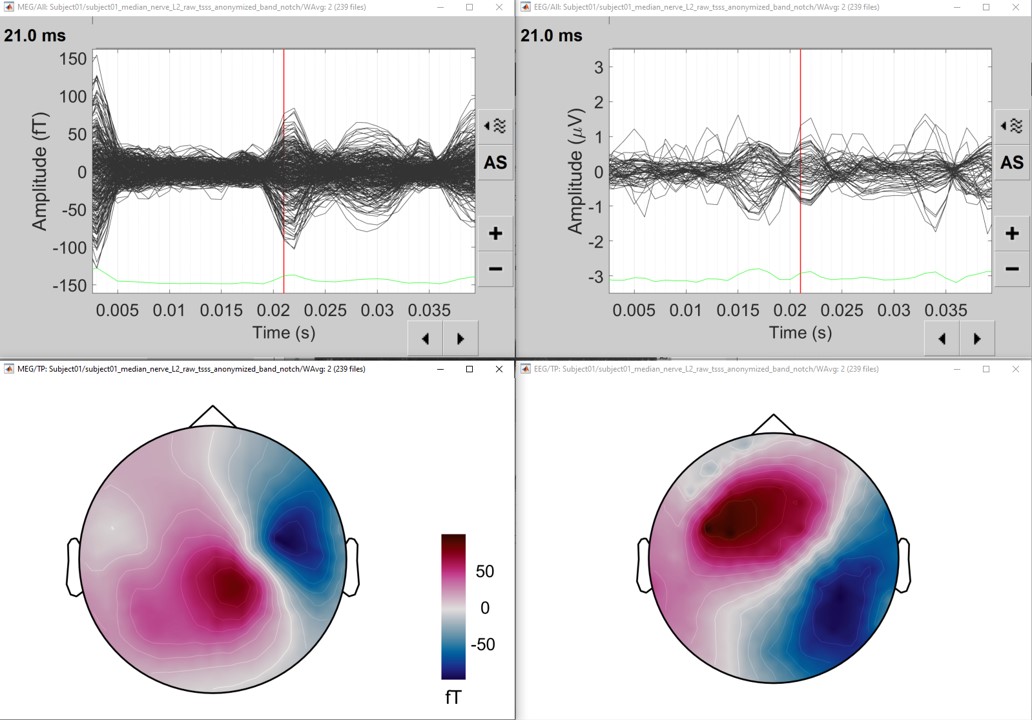
- At the selected time (21ms), an ERP is visible with a nice dipolar pattern on the sensors, both for the EEG and MEG.
Forward model
We are going to use the realistic FEM model previously generated from the MRI. Go to the "Anatomy" view, and make sure that the FEM head model is highlighted in green color (it should be the case if you have only one model). You may also highlight the cortex to use for the computation (select the cortex_15002V).
You can compute the forward model both for EEG and MEG simultaneously, however, using the high mesh resolution model we recommend to compute separately the head model for each modality (EEG and then MEG). The time required for EEG is aound one hour for ~70 channels, for the MEG with 306 sensors it can take up to 4 hours or more (with the integrations points).
The EEG/MEG FEM computation depends on the computation of the FEM transfer matrix, which is related to the resolution of the FEM head mesh (number of vertices) and the number of sensors. In most of the case, the number of EEG sensors is lower than the number of MEG sensors. Furthermore, internally the MEG sensors modeling uses the integrations points, which increase the number of computation points (~ multiplied by 4 for the magnetometers and by 8 for the gradiometers). Therefore the MEG requires more time than the EEG.
To reduce the MEG computation time, there are some tips :
Use only the inners tissues (wm, gm, and CSF) ==> reduce the number of vertices
Do not use the integration points ==> reduce the number of virtual sensors
- These parameters can be tuned from the DUNEuro options panel (see the advanced panel)
EEG with DTI tensors
- Go back to the "Functional data" view
Right-click on the channel file (folder with imported epochs) > Compute head model
Select EEG only > DUNEuro FEM, and set the comment to "DUNEuro FEM EEG DTI tensors"
- On the DUNEuro options panel: click on "Show details" and set the options as illustrated below
Click OK. Depending on the performance of your computer, the FEM EEG head modeling can last between 30min to 3hrs. Once the computation is done, a new node with the name "DUNEuro FEM EEG DTI tensors" appears in the Brainstorm database navigator, which can be used for the source localization process.
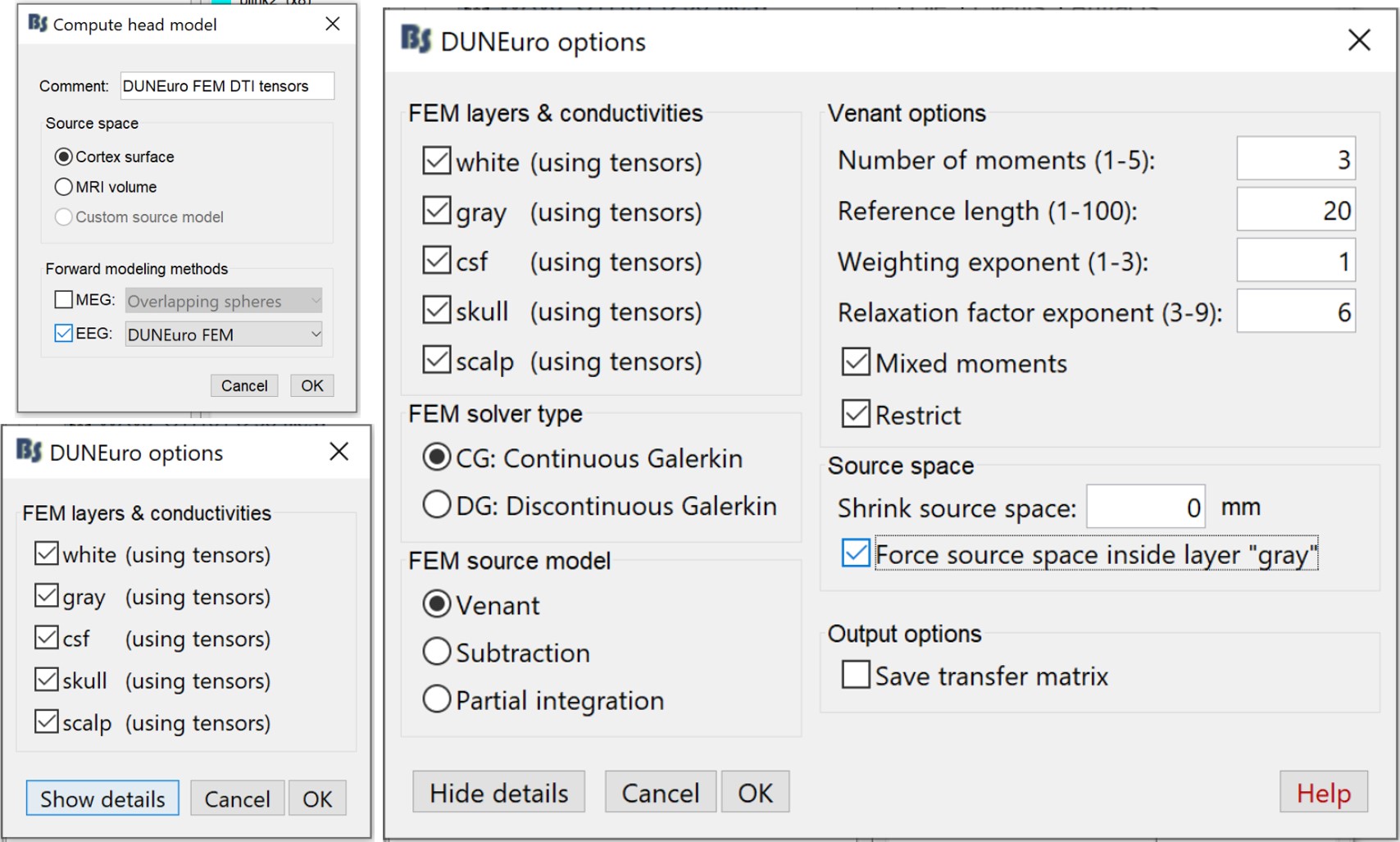
MEG with DTI tensors
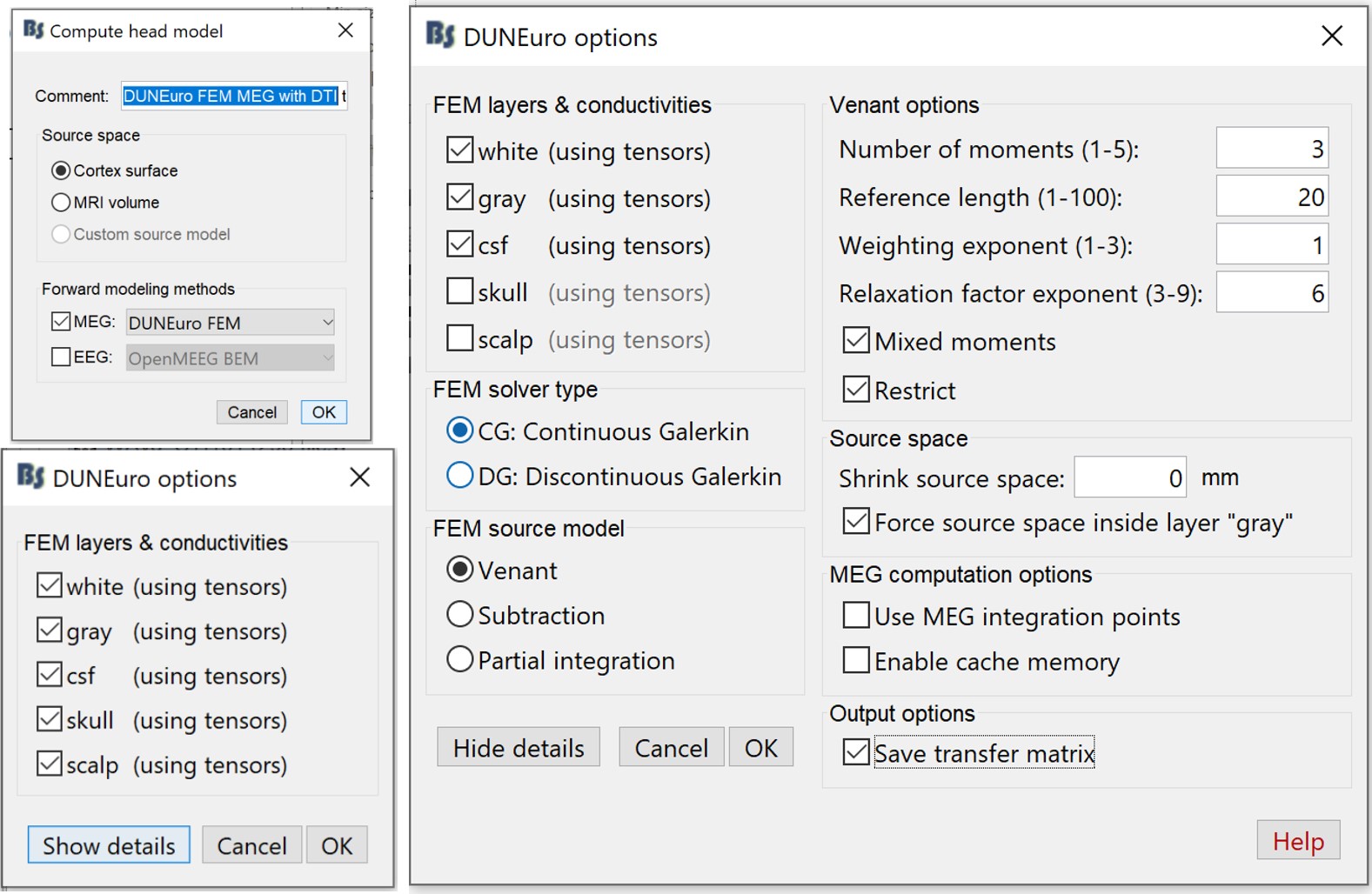
Right-click on the channel file > Compute head model
Select MEG only > DUNEuro FEM, and set the comment to "DUNEuro FEM MEG DTI tensors"
- On the DUNEuro options panel: click on "Show details" and set the options as illustrated below. To reduce the computation time, we select only the inner layers (white, gray and CSF).
Click OK. Depending on the performance of your computer, the FEM MEG head modeling can last between 1hr to 4hrs. Once the computation is done, a new node with the name "DUNEuro FEM MEG DTI tensors" appears in the Brainstorm database navigator, which can be used for the source localization process.
EEG with isotropic conductivity
If the DTIs are not available, it is possible to use the isotropic conductivities instead. In this tutorial, we will compute another forward model in order to compare the sources obtained with anisotropic or isotropic conductivities.
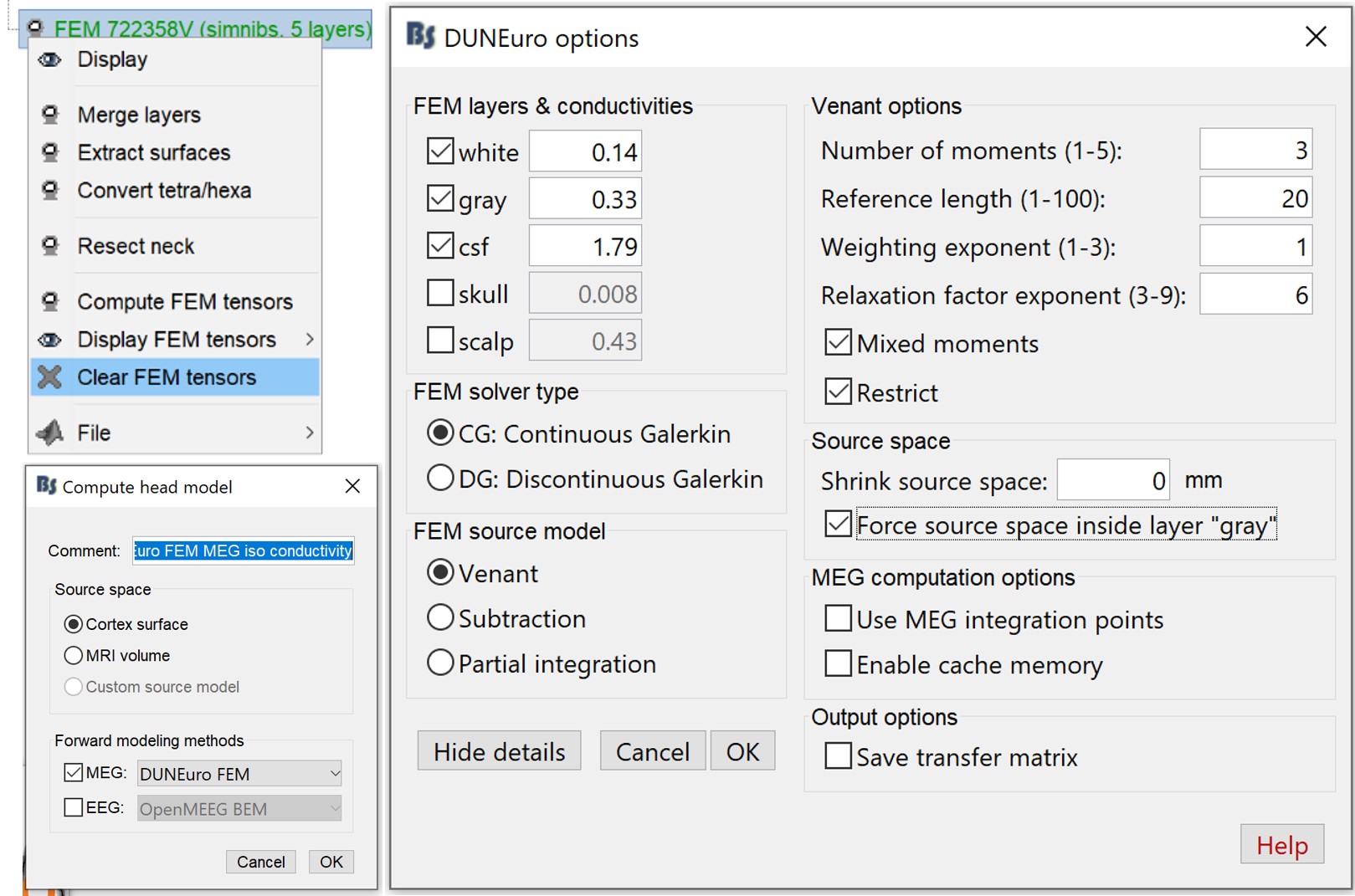
First you need to make sure that the tensors are not computed. If you have the tensors already computed, you need to clear these values. Go to the "Anatomy" view, right-click on the FEM mesh > Clear FEM tensors.
Go back to "Functional data", right-click on channel file > Compute head model. In this case, you can change the isotropic conductivity for each tissue. Set the options as in the figure below.
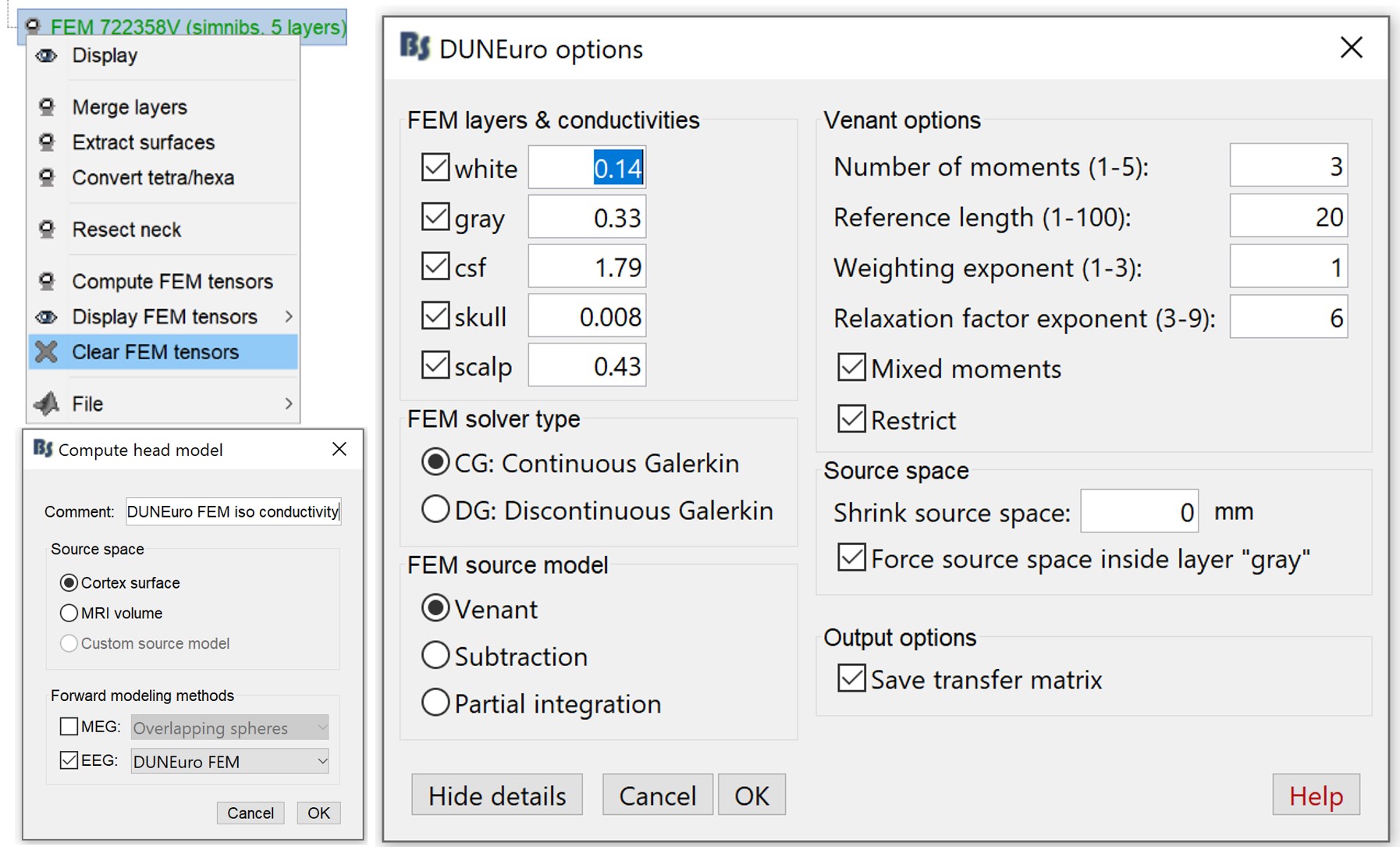
MEG with isotropic conductivity
- Same as before, first you need to make sure that the tensors are not computed.
Right-click on the channel file > Compute head model, and set the value as in the figure below. In this example keep only the inner tissues (white, gray and CSF).
Source estimation
Noise covariance matrix
- We will use the baseline of the single epochs to calculate the noise covariance matrix.
Right-click on the Trigger01 epochs group > Noise covariance > Compute from recordings.
Enter the same baseline interval we used for removing the DC offset: [-100, -10] ms 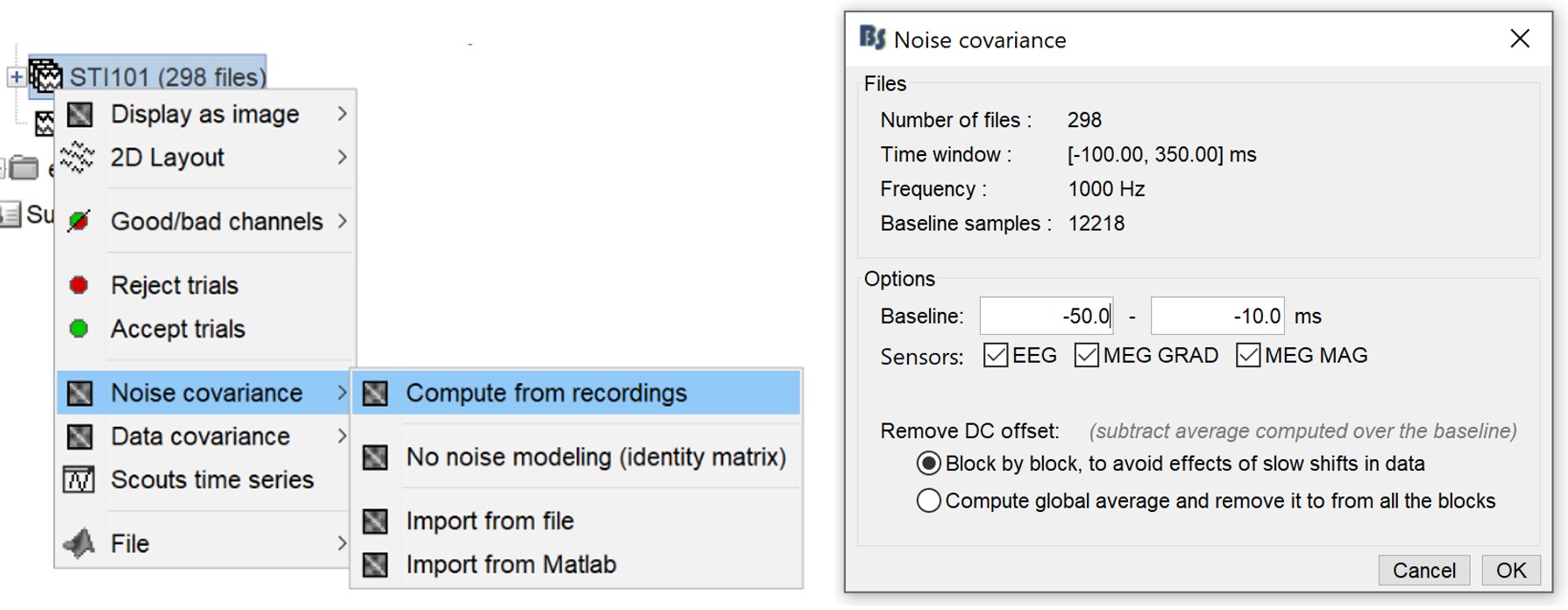
Inverse model
Right-click on the head model > Compute sources.
Select "Dipole modeling option" and EEG.
Repeat the same operation for MEG and make sure to select the appropriate head model at each step.
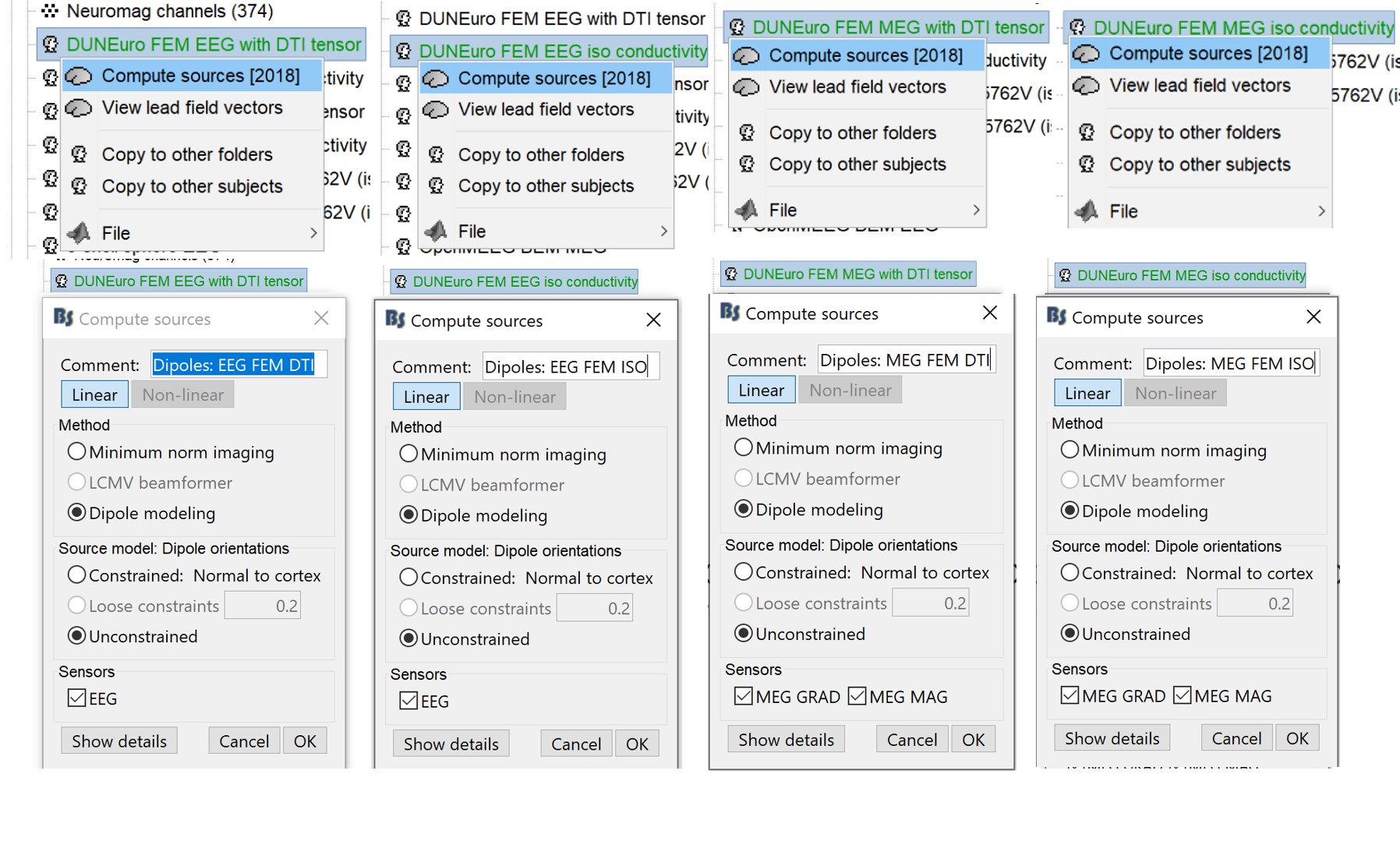
It is better to study separately the two modalities because the method for combining MEG and EEG are not working well yet within Brainstorm.
This operation creates a shared inversion kernel and one source link for each block of recordings in the folder.
Drag and drop the Dipoles kernels in the Process table and select the process > Run> Dipole Scanning>
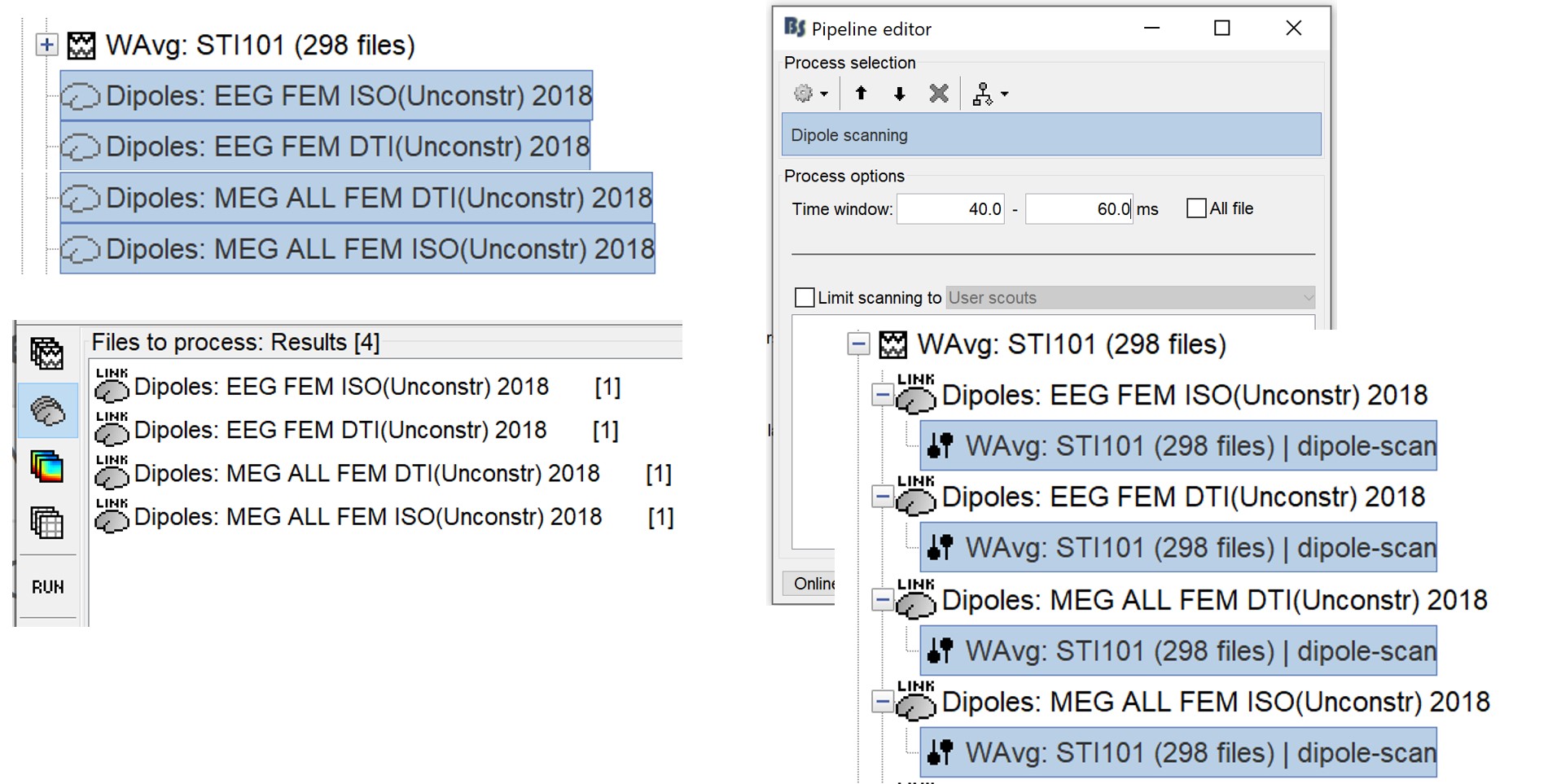
If you are not familiar with those concepts, please refer to the Source estimation tutorial.
Once the dipoles are computed, we display both EEG and MEG separately in the following figures. We merge the MEG ISO and the MEG DTI dipoles as well as the EEG dipoles.
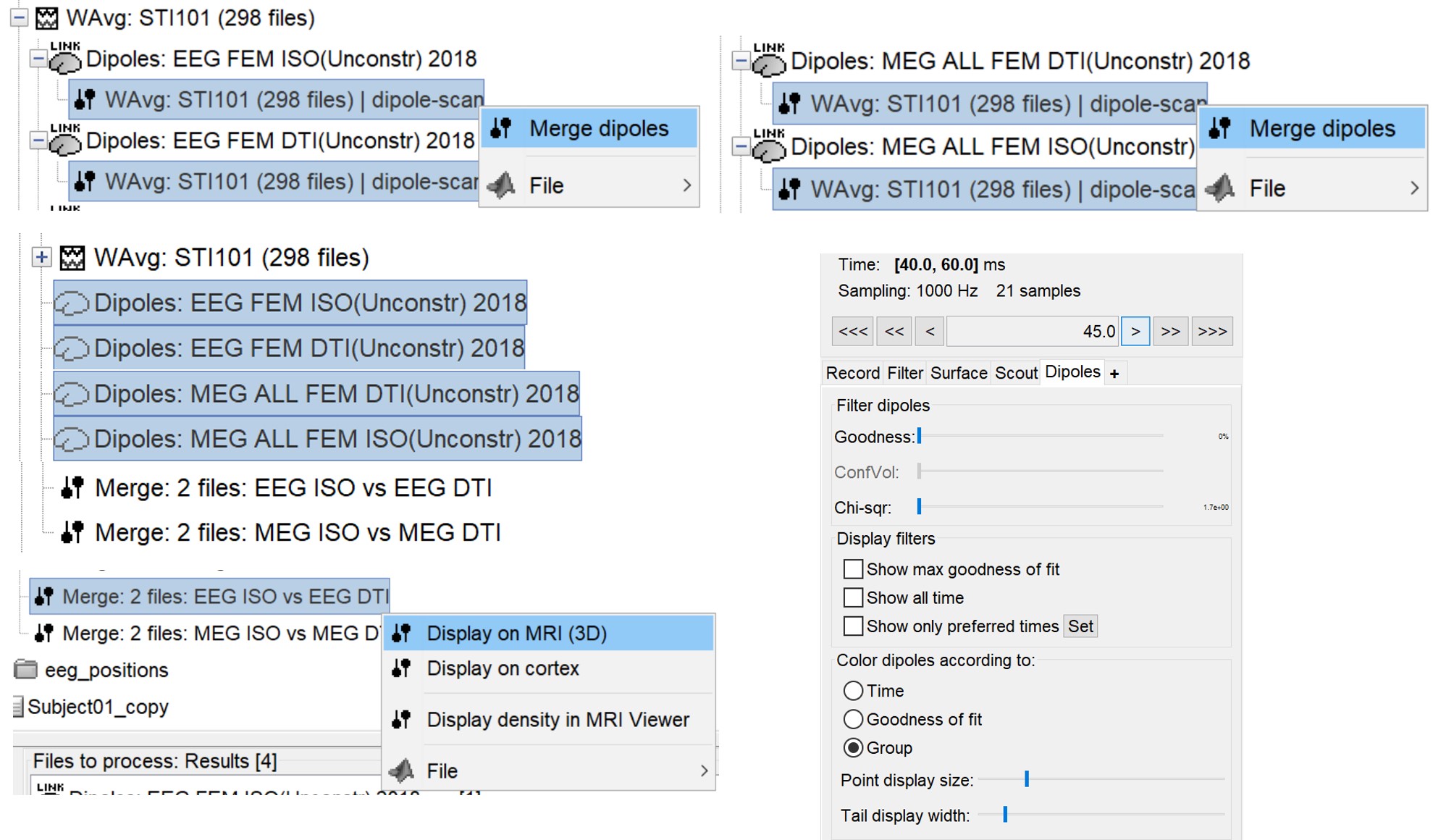
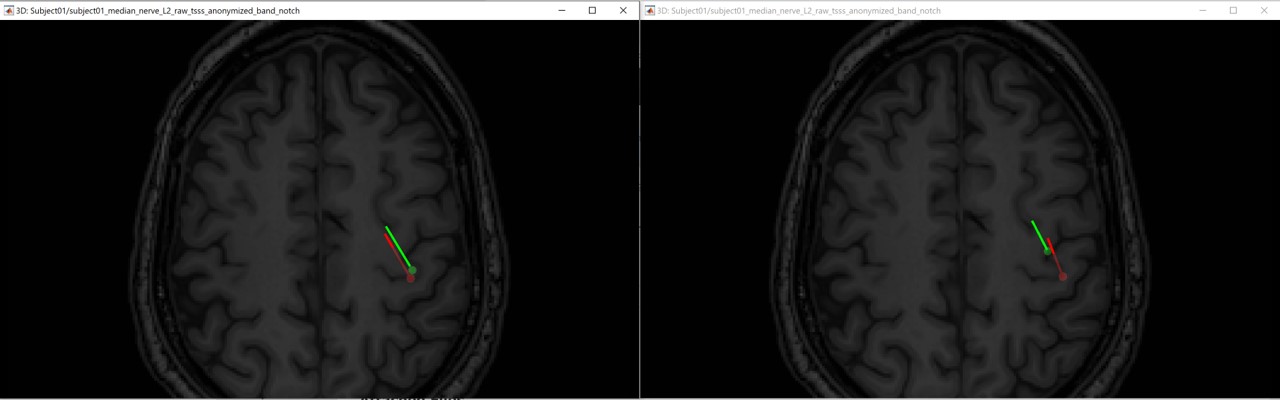
P20/N20 source localization
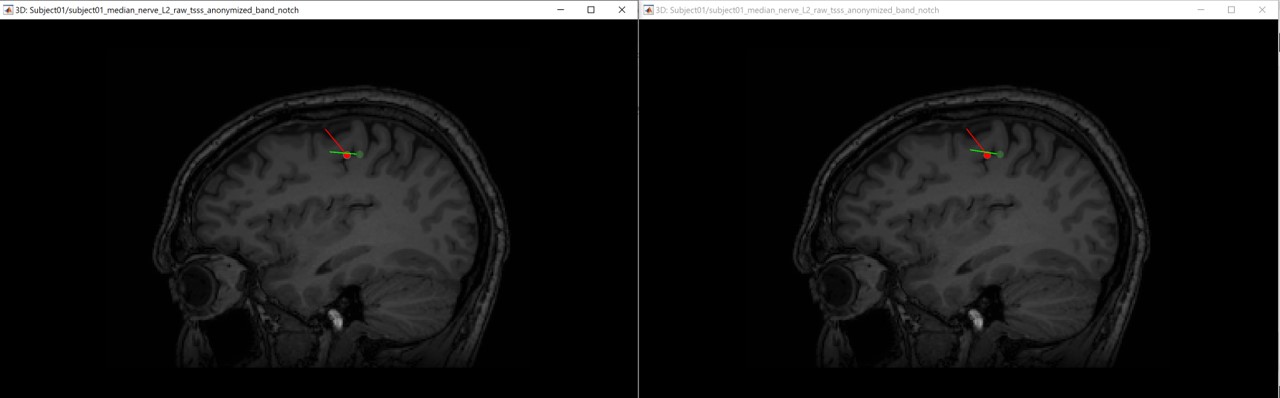
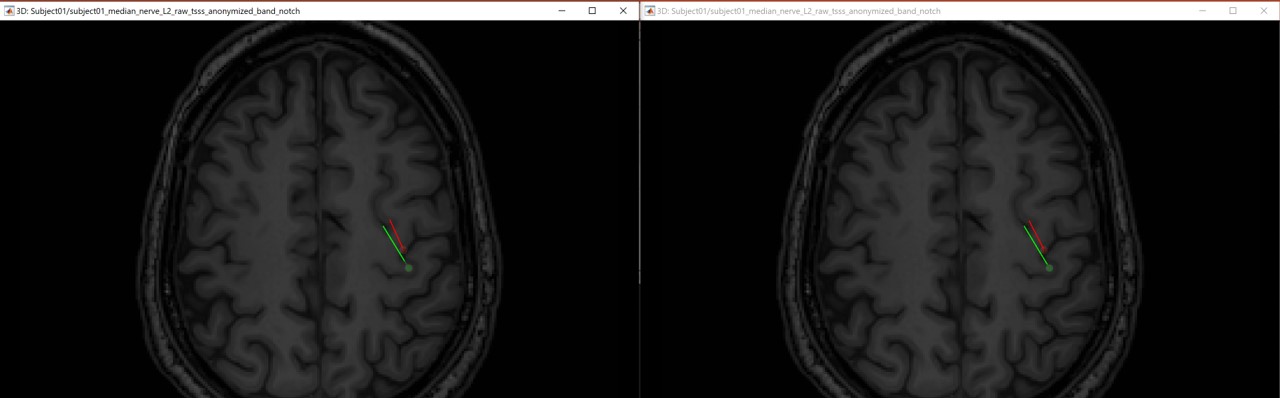
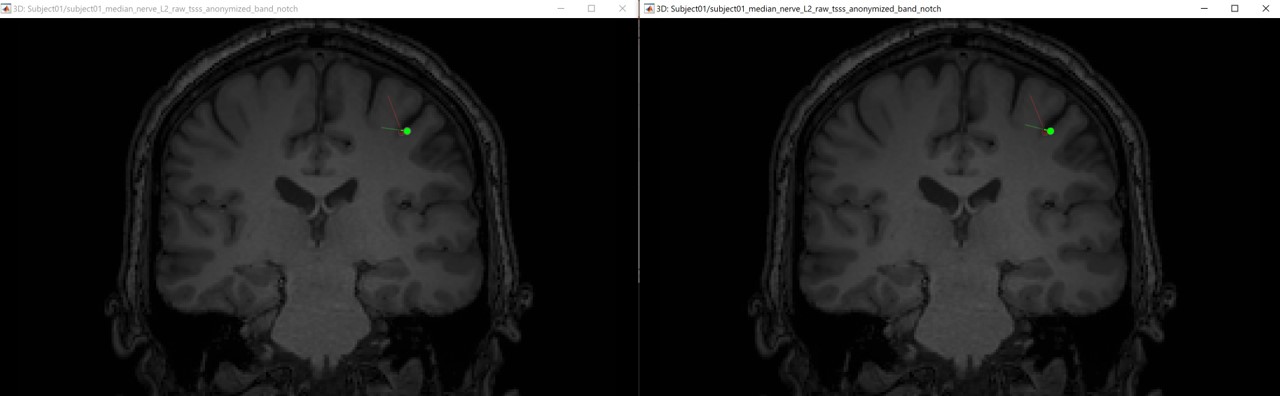
The following figures (left EEG, right MEG), show the localization of the dipoles on the MRI at the 21ms. The ISO model is colored in greeen wherease the DTI model is colored in red.
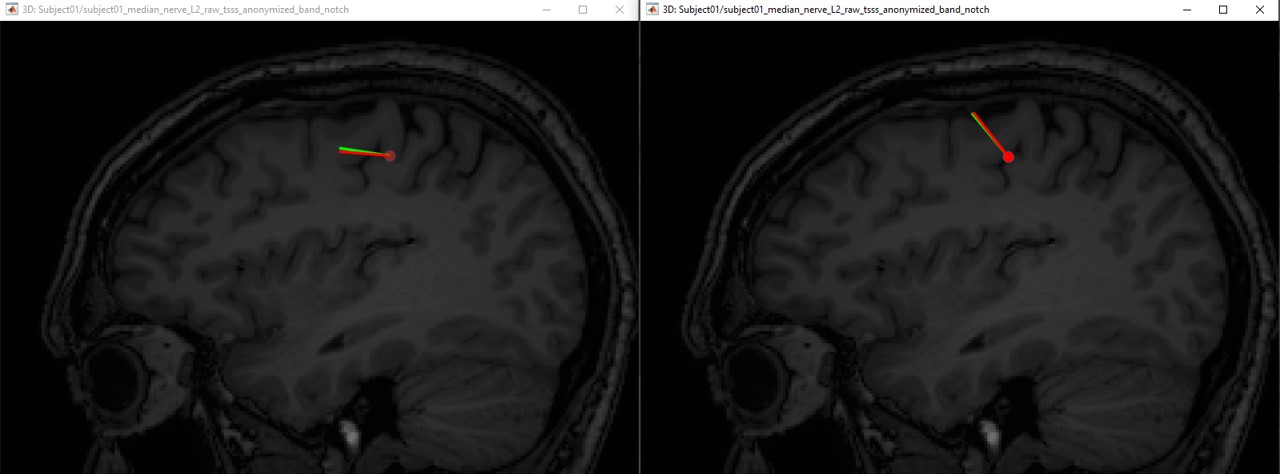
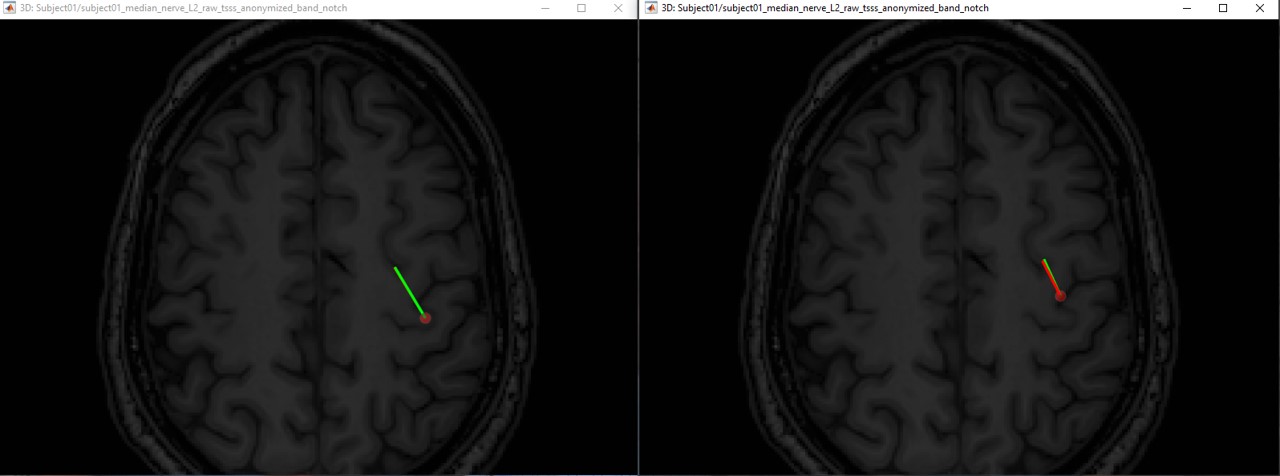
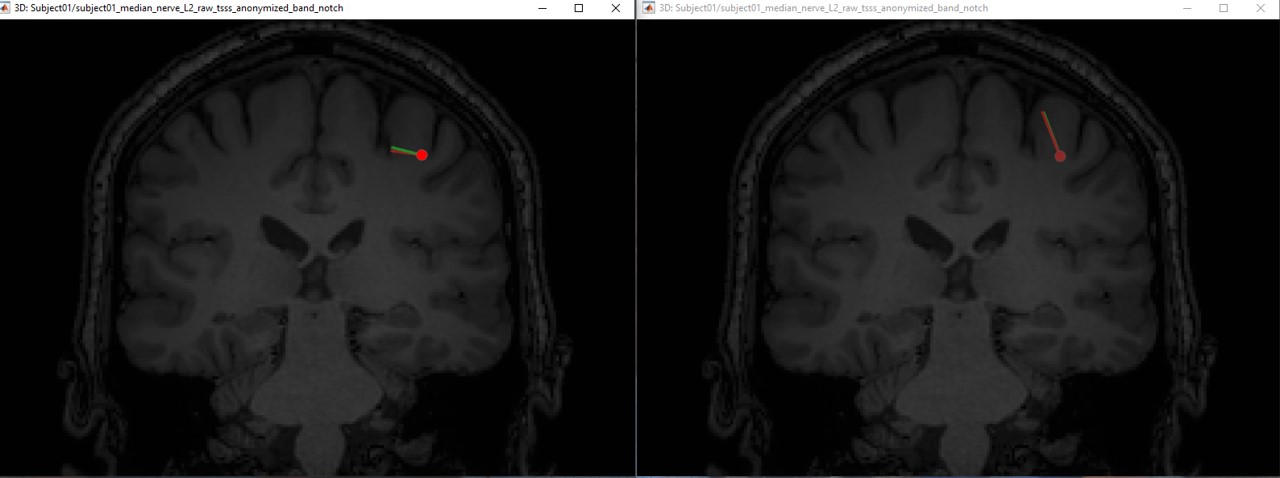
In this experiment, the anisotropy does not show a significant effect on the source localization, whereas it shows a slich difference in the orientation.
When we compare between the EEG and the MEG, there is a difference of 20mm between the localization between the MEG and the EEG as well as a difference in the orientation in the coronal view.
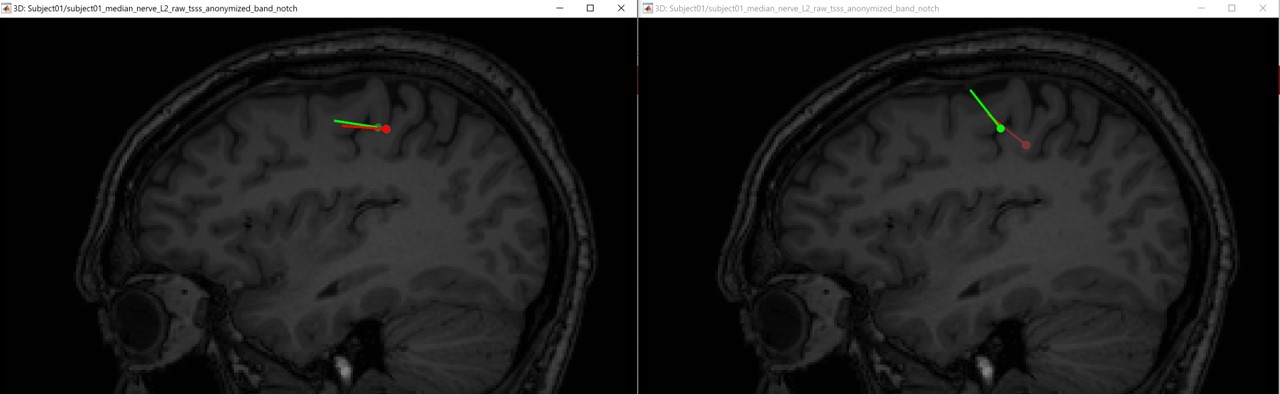
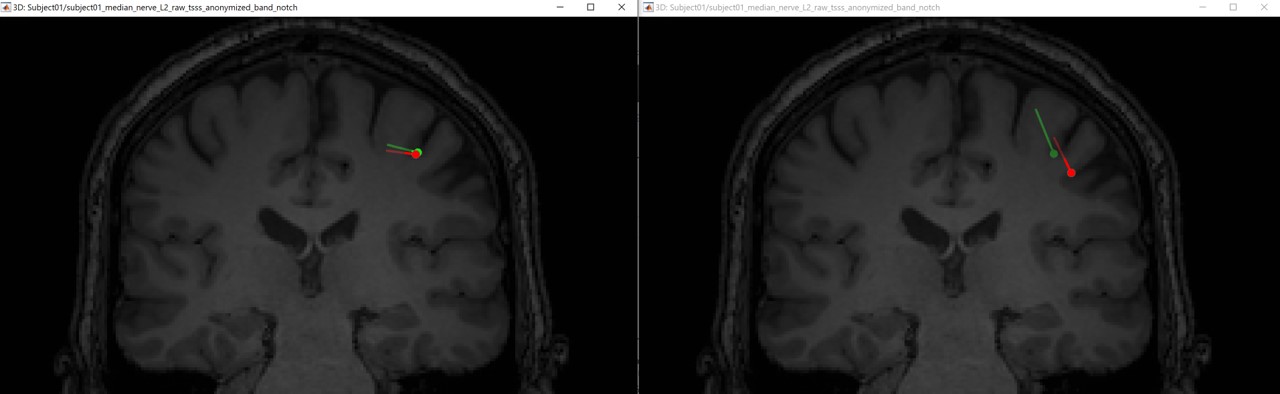
If we compare the difference between the EEG and MEG dipoles, the following figures (left anisotropy, right isotropic), show the localization of the dipoles on the MRI at the 21ms. The EEG dipole is colored in green wherease the MEG dipole is colored in red.
Aniso: Eeg [-7.5 -35.7 89.8] vs Meg [-0.2 -33.6 88.6] ==> distance = 7.7mm
Iso: Eeg [-7.5 -35.7 89.8] vs Meg [-0.2 -33.6 88.6] ==> distance = 7.7mm
In both modalities, the dipoles are located exactely in the same positions, and there is 7.7mm between the two dipoles. Further more, we notice a difference on the orientation on the coronal and sagital views
Comparaison with BEM for EEG and with OS for the MEG
In order to compare qualitatively the results of the FEM models, we will use the previous methods already implemented in Brainstorm.
For the EEG: compute the forward using the BEM For the MEG: compute the forward using overlapping spheres,
For all the methods, we will perform the dipole scanning as done previously but with the new forward models, the results are shown in the following figures:
In the following figures, left is the dipoles computed from the EEG (BEM in red and FEM in green), right are the dipoles computed from the MEG (OS in red and FEM in green)
EEG: fem [-7.5 -35.7 89.8] ; bem [-10.7 -35.0 89.5] ==> 3.3mm
MEG: fem [-0.2 -33.6 88.6] ; os [-10.7 -39.3 83.2] ==> 13.1 mm
As expeced, the slight difference on the localization can be explained by the difference on the head shape, the conductivity values as well as the resolution method.
Resect the neck and the bottom part of the head model
The FEM computation can be time and memory-consuming, on way to reduce this issue is to remove the part of the FEM mesh that is not required.
In some cases where the field of the MRI is large, you can have the mesh of the neck and even the shoulders. In Brainstorm, it is possible to remove these parts. You can this by following these steps : right-click on the FEM-mesh => Resect Neck ==> keep the default value ==> Ok
Once the process is finished, a new FEM head model appears in the database with the same name and a prefix "resect" which is the resected head. The following figure shows the model before and after re-section.
Note that the initial model has 743828 nodes and 4079587 and the resected model has 613955 nodes and 3400957 elements.
Scripting
This section is under developement
DUNEuro advanced options
This section is under development
A set of advanced options are made available and can be easily changed. Here a short explanation is given for each option.
FEM layers & conductivities: same explanation as in the previous section. Moreover, in the case where the conductivity tensor are previously computed, Brainstorm detects these tensors and load them. In this case, the users can't change the conductivities scalar values, since they are not used. The following is displayed panel is displayed.
FEM solver type:
- CG or Continuous Galerkin: This is the standard Lagrangian method.
- DC or the Discontinuous Galerkin:
In this version, only the Fitted FEM approaches are integrated, that require the FEM mesh of the head model. Unfitted methods are also available within DUNEuro, and will be integrated soon in Brainstorm.
FEM source model: The list of the available source are
- Venant
- Subtraction
- Partial Integration
For more information about these methods, users can check this thesis (Vorwek thesis).
- Source space
- Shrink source space: the location of dipoles are moved inward by the specified value in this field(in mm).
- Force source space: this is required in the case where the dipoles are not within the GM matter.
- Outputs options
- Save transfer matrix:
All these parameters are stored and passed to the DUNEuro as a text file. This file is the main interface that passes the parameters from Brainstorm to DUNEuro. More details about the integration can be found in these links:
GitHub repository for the Brainstorm-DUNEuro compilation and integration
GitHub repository for the matlab-duneuro interface
- Brainstorm-DUNEuro integration discussions:
Troubleshooting and solution(TODO)
Dipoles outside of the grid for the MEG: remove this dipole or replace it with its spatial neighbors (not the neighbor in the list) ==> not easy to handle write small tuto?