Tutorial: Process functional NIRS data
|
|
Authors: Thomas Vincent
Prerequisite:
Presentation of the experiment
- Finger tapping task: 10 stimulation blocks of 30 seconds each, with rest periods of ~30 seconds
One subject, one NIRS acquisition run of 12 minutes at 10Hz
- 4 sources and 12 detectors (+ 4 proximity channels) placed above the right motor region
- Two wavelengths: 690nm and 830nm
MRI anatomy 3T from
 scanner type
scanner type
Extract stimulation events
During the experiment, the stimulation paradigm were run under matlab and sent triggers through the parallel port to the acquistion device. These stimulation events are then stored as a box signal in channel AUX2: values above a certain threshold indicate a stimulation block.
To transform this signal into Brainstorm events, drag and drop the NIRS data "S01_Block_FO_LH_Run01" in the Brainstorm process window. Click on "Run" and select Process "Events -> Detect events above threshold".
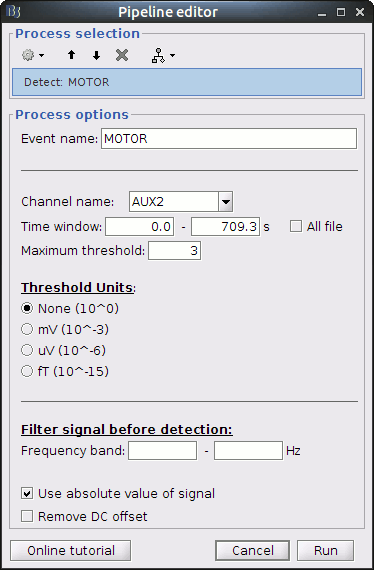
Use the following parameters:
- set "Event name" to "MOTOR"
- select "Channel name": "AUX2"
- set "Maximum threshold" to 3
- Check "Use absolute value of signal"
Then run the process.
To view the results, right-click on "Link to raw file" under "S01_Block_FO_LH_Run01" then "NIRS -> Display time series".
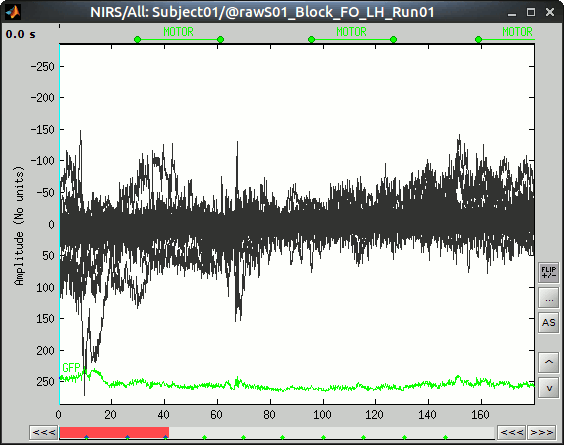
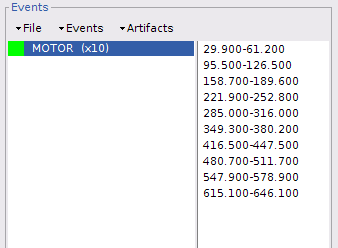
The "MOTOR" event group has been created, by 10 events, each lasting 30 sec. Events are shown in green on the top of the plot.
Bad channel tagging
NIRS measurement are heterogeneous (long distance measurements, movements, occlusion by hair) and the signal in several channels might not be analyzable. A first pre-processing step hence consists in removing those channels.
The following criterions may be applied to reject channels:
- some values are negative
- signal is flat (variance close to 0)
- signal has too many flat segments
Drag and drop the NIRS data "S01_Block_FO_LH_Run01" in the Brainstorm process window. Click on "Run" and select Process "NIRSTORM -> Tag bad channels".
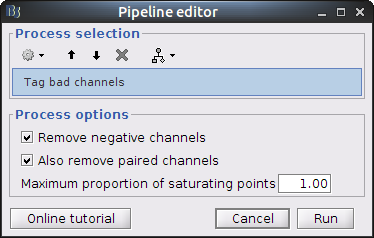
- Remove negative channels: tag a channel as bad if it has a least one negative value. This is important for the quantification of delta [Hb] which cannot be applied if there are negative values.
- Maximum proportion of saturating point: a saturating point has a value equals to the maximum of the signal. The default is at 1: remove only flat signals. If one wants to also keep flat channels, set the value to at least 1.01.
This process is performed "in place": the channel flags of the given data are modified. To view the result, right-click on "S01_Block_FO_LH_Run01 > Link to raw file" then "Good/Bad Channels > View all bad channels" or "Edit good/bad channels".
Delta Hb - Modified Beer-Lambert Law
This process computes variations of concentration of oxy-hemoglobin (HbO), deoxy-hemoglobin (HbR) and total hemoglobin (HbT) from the measured light intensity time courses at different wavelengths.
Note that the channel definition will differ from the raw data. Previously there was one channel per wavelength, now there will be one channel per Hb type (HbO, HbR or HbT). The total number of channels may change.
For a given pair, the formula used is:
delta_hb = d-1 * eps-1 * -log(I / I_ref) / ppf
where:
delta_hb is the 3 x nb_samples matrix of delta [Hb],
d is the distance between the pair optodes,
eps is the 3 x nb_wavelengths matrix of Hb extinction coefficients,
I is the input light intensity,
I_ref is a reference light intensity,
ppf is the partial light path correction factor.
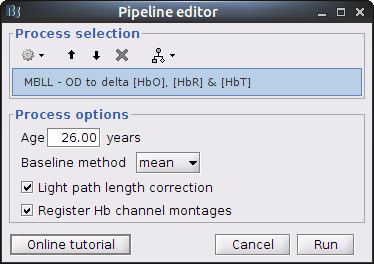
Process parameters:
- Age: age of the subject, used to correct for partial light path length
Baseline method: mean or median. Method to compute the reference intensity (I_ref) against which to compute variations.
Light path length correction: flag to actually correct for light scattering. If unchecked, then ppf=1
Register Hb channel montages: register one montage per paired channel group. This will create a list of montage to help visualizing the different Hb time-courses together.
 make ref to visu
make ref to visu