
Neuromaps plugin
note: this tutorial page is currently under construction.
The Neuromaps Brainstorm plugin integrates curated annotations and tools to further expand the accessibility and inclusivity of brain-mapping tools, as part of an Open Science initiative. We extend these pioneering tools to the MATLAB environment for users without any prior computer programming experience, providing an intuitive graphic user interface.
The present tutorial will demonstrate the plugin’s functionality within the Brainstorm interface; for a detailed breakdown of the algorithm, please refer to the neuromaps plugin Github.
Contents
- Introduction
- Plugin Key Features
- Installing and Running the Neuromaps Plugin
- Importing the Brain Annotations
- Visualizing and Accessing Annotation Parameters
- Statistical Analyses for Significance Testing
- On the Hard Drive
- Applications of Neuromaps for Integrative Research
- Acknowledgements
- Contributing
Introduction
Neuromaps is a toolbox designed for assessing, transforming, and analyzing structural and functional brain annotations (Markello, Hansen et al., 2022). It marks a significant stride towards integrative analytics in multimodal, multiscale neuroscience by bridging the gap between various neuroimaging modalities and proposing analytical tools to establish connections among different brain features. The toolbox comprises a curated repository of brain maps in their native space, methods for generating transformations across multiple coordinate systems, and offers a systematic workflow for comprehensive structural and functional annotation enrichment analysis of the human brain.
For the first iteration of the implementation, we focused on the neurotransmitter receptors and transporters. Thirty different maps from the neuromaps toolbox were selected, covering nine different neurotransmitter systems: dopamine, norepinephrine, serotonin, acetylcholine, glutamate, GABA, histamine, cannabinoid, and opioid. These maps are sourced from open-access repositories, addressing the need for a comprehensive tool that integrates standardized analytic workflows for both surface and volumetric data.
Plugin Key Features
- Two different coordinate systems:
FsAverage for surface maps- the default system used by the FreeSurfer software.
MNI152 for volumetric maps- 152 normative MRI scans developed by the Montreal Neurological Institute (to be added soon).
Flexible framework designed to accommodate future expansions and updates.
Repository of precomputed annotations sourced from the published literature for both volumetric and surface systems, ensuring accessibility and ease of use.
All the information for these maps, including the appropriate citations to use can be found in this spreadsheet.
To obtain the surfaces, the original maps offered in MNI152 space were transformed to FsAverage using the registration fusion framework proposed in neuromaps (Buckner et al., 2011; Wu et al., 2018).
Correlation processes that account for spatial autocorrelation.
Installing and Running the Neuromaps Plugin
From the main window go to Plugins > Anatomy > neuromaps > Install.
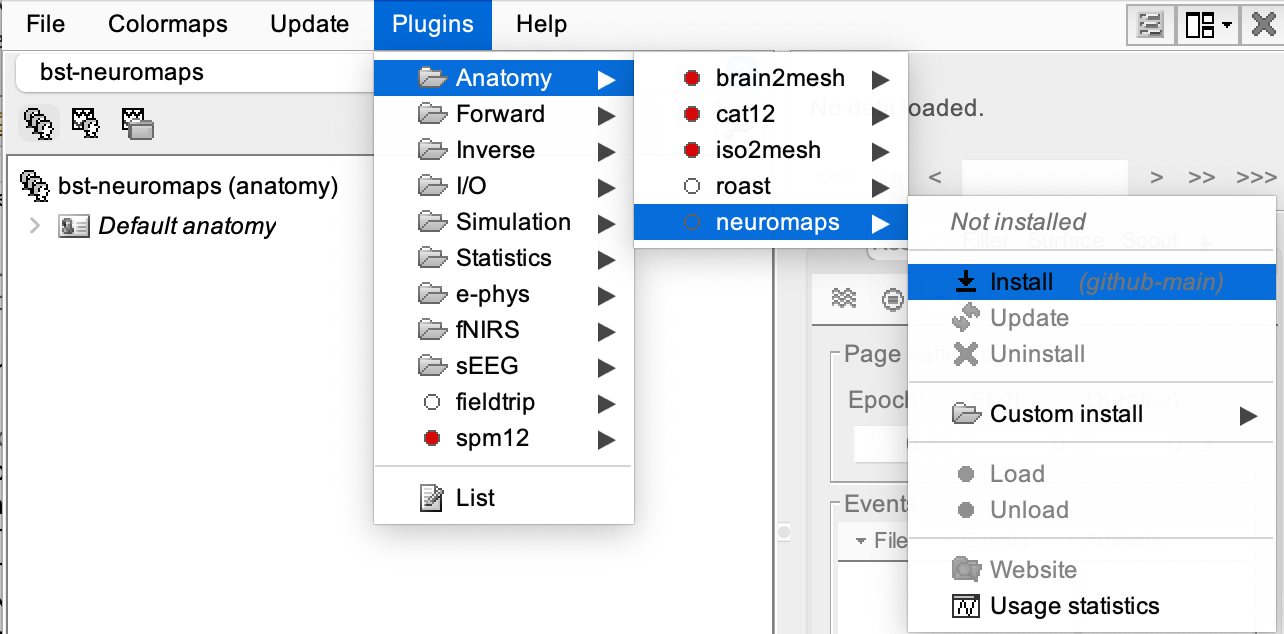
A message will appear saying, "Plugin 'Neuromaps' is not installed on your computer. Download the latest version of Neuromaps now?" click 'Yes'.
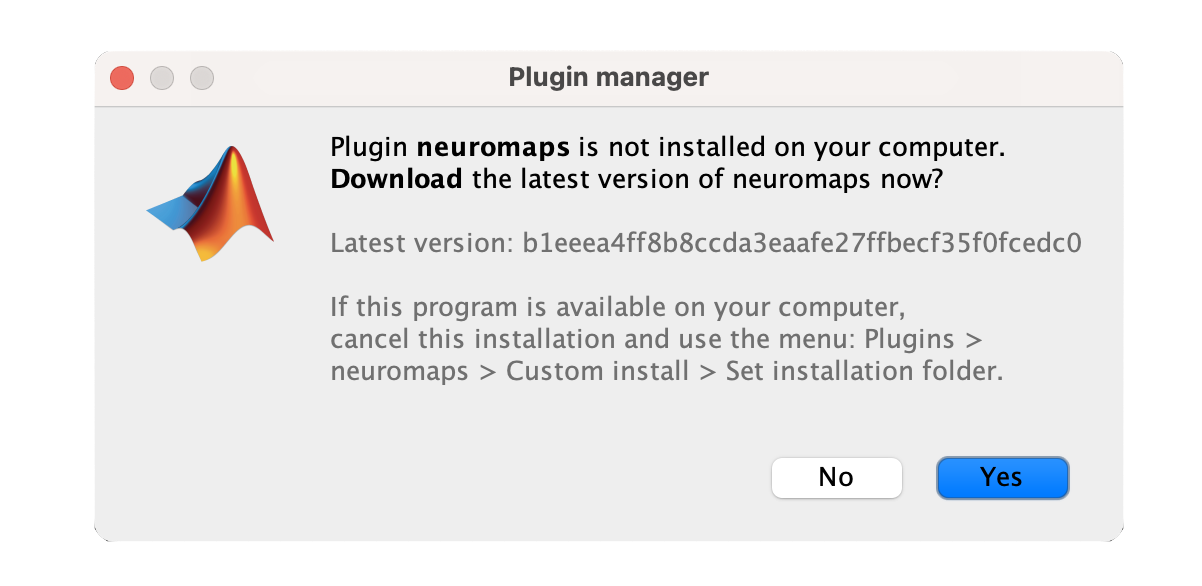
The plugin will be downloaded and installed automatically. Once installed, you will see a confirmation message.
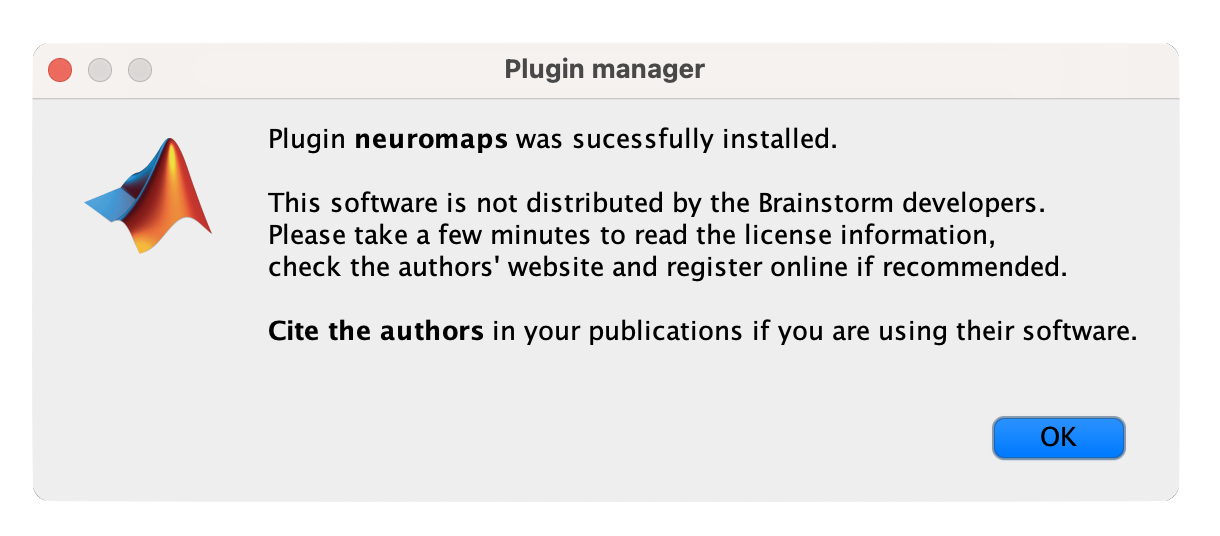
- The README file will appear; after thoroughly reviewing it, click on the "I agree" button to confirm your acceptance of the plugin's terms and conditions.
By following these steps, you will successfully install the Neuromaps plugin.
Importing the Brain Annotations
Click on the [Run] button at the bottom-left corner of the Process1 tab.
When the Pipeline editor window appears, click on the first button in the toolbar under 'Process selection' (the one with a gear icon). It will show you a list of processes.
Go to Sources > Fetch brain annotations from Neuromaps.
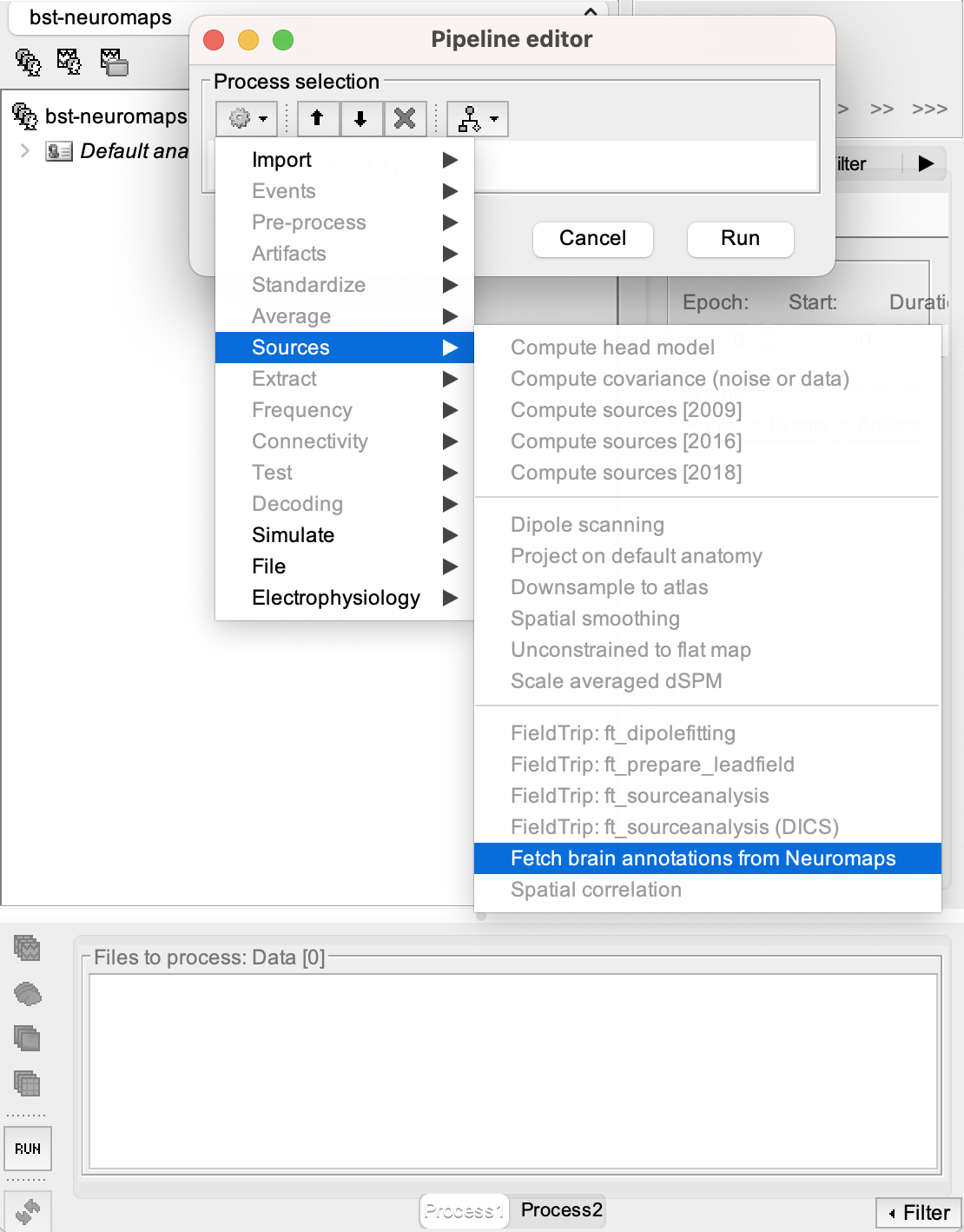
- You will be able to switch between 'Surface' or 'Volume' for the ‘Source space’ process option.
The brain annotations will be listed right under, in the name format '[neurotransmitter]:[subtype]_[tracer]_[source]_[size]_[age]' (e.g., 'Acetylcholine: VaCht_feobv_aghourian2017_N18_Age67').
Select all the maps you want to import and click Run.
Visualizing and Accessing Annotation Parameters
Once you have fetched the brain annotations, a new 'Neuromaps' node which contains the FSAverage anatomy will appear in the Anatomy view.
All the imported brain annotations will be visible in the Functional data (sorted by subjects) view.
To visualize the surfaces, you can either double-click on the file or right-click > Cortical activations > Display on cortex.
Visualization capabilities
Implementing neuromaps directly into Brainstorm further enhances the overall user experience by providing greater flexibility and control over the visualization and analysis of brain annotations. Brainstorm's advanced functionalities allow users to effortlessly rotate the brain annotations, zoom in and out, reposition them, obtain the coordinates for any point, take snapshots, adjust the transparency, and more.
The ‘Surface’ tab of the Brainstorm window contains buttons and sliders to control the display of these surfaces. Additional relevant functionalities, as outlined in Tutorial 3: Display the Anatomy, include:
Smooth: Inflates the surface to make all the parts of the cortex envelope visible.This is just a display option, it does not actually modify the surface.
Resect: The sliders and the buttons Left/Right/Struct at the bottom of the panel allow you to cut the surface or reorganize the anatomical structures in various ways.
Edge: Display the faces of the surface tesselation.
Colormap
The default colormap upon opening the brain annotation is royal_gramma, but you have the option to switch to other sequential, diverging, or rainbow colormaps. Simply right-click on the brain annotation window, navigate to Colormap: Sources > Colormap.
If you modify a colormap, the changes will be applied to all the figures, saved in your user preferences and available the next time you start Brainstorm. If you wish to undo the changes and reset the colormap to its default values, click on Restore defaults or double-click on the color bar.
You also have the opportunity to customize the brightness, contrast, and decide the color mapping, among other settings. For a comprehensive list of available options with detailed instructions, please refer to Tutorial 18: Colormaps.
Creating Weighted-Averaged Annotations
Neurotransmitter annotations that have the same tracer (e.g., Serotonin: 5-HT1b_p943_gallezot2010_N23_Age29 and Serotonin: 5-HT1b_p943_savli2012_N23_Age29 have the same p943 tracer) can be combined to obtain a larger sample size for your analysis. To accomplish this, you can perform a weighted average. In the same way you selected the fetching process:
- Drag and drop all the relevant brain annotation files into the processing pipeline (Process1). In this example, we will select the two 5-HT1b files mentioned.
Click on the [Run] button at the bottom-left corner of the Process1 tab.
Once the Pipeline editor window appears, click on the first button in the toolbar under 'Process selection' (the one with a gear icon).
Select the process Average > Average files.
Under 'Group files,' select Everything and for 'Function,' Arithmetic average: mean(x).
Check the 'Weighted average' box, as shown below.
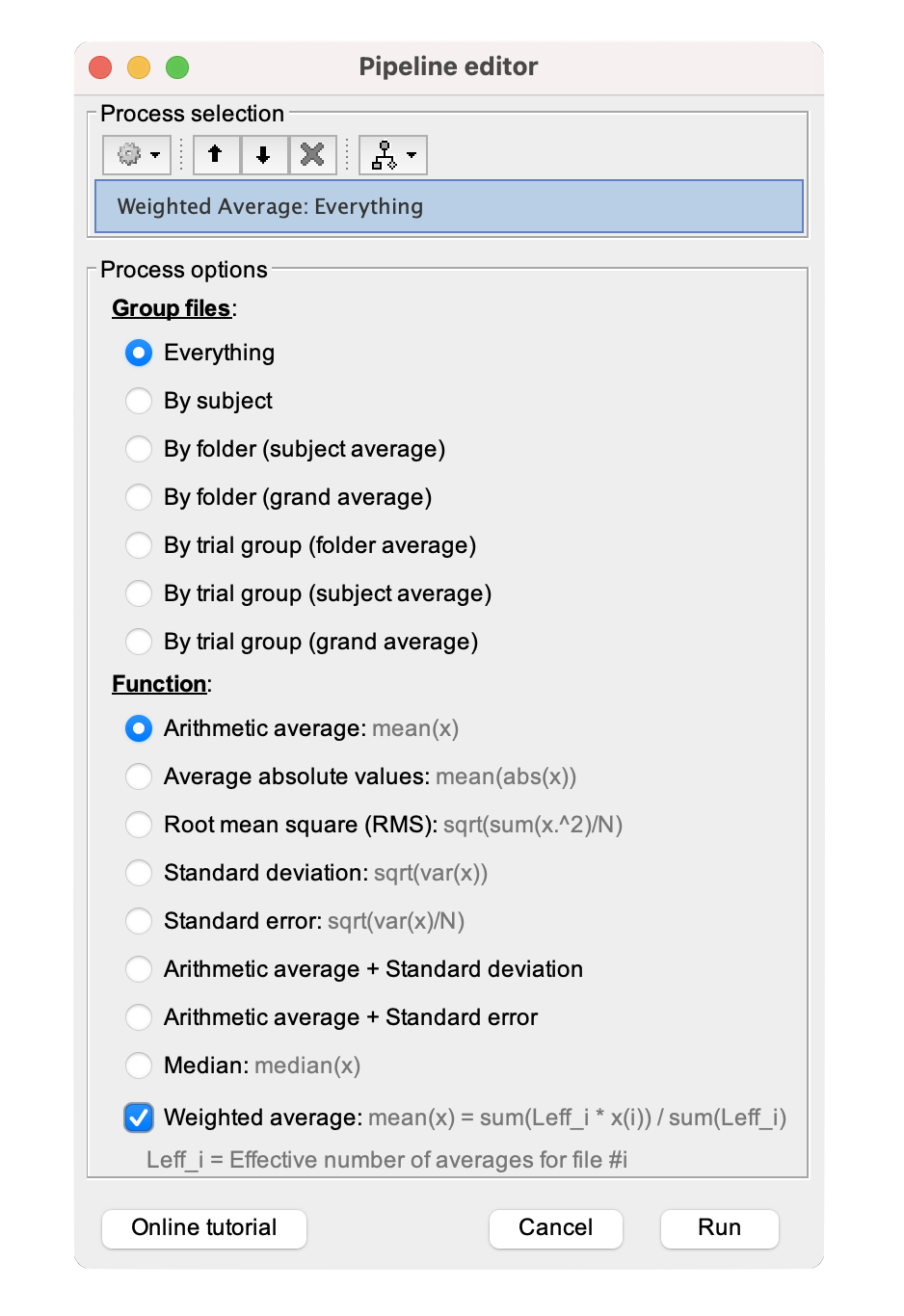
Click Run. A new file 'WAvg: 2 files' will be added under the same neurotransmitter folder.
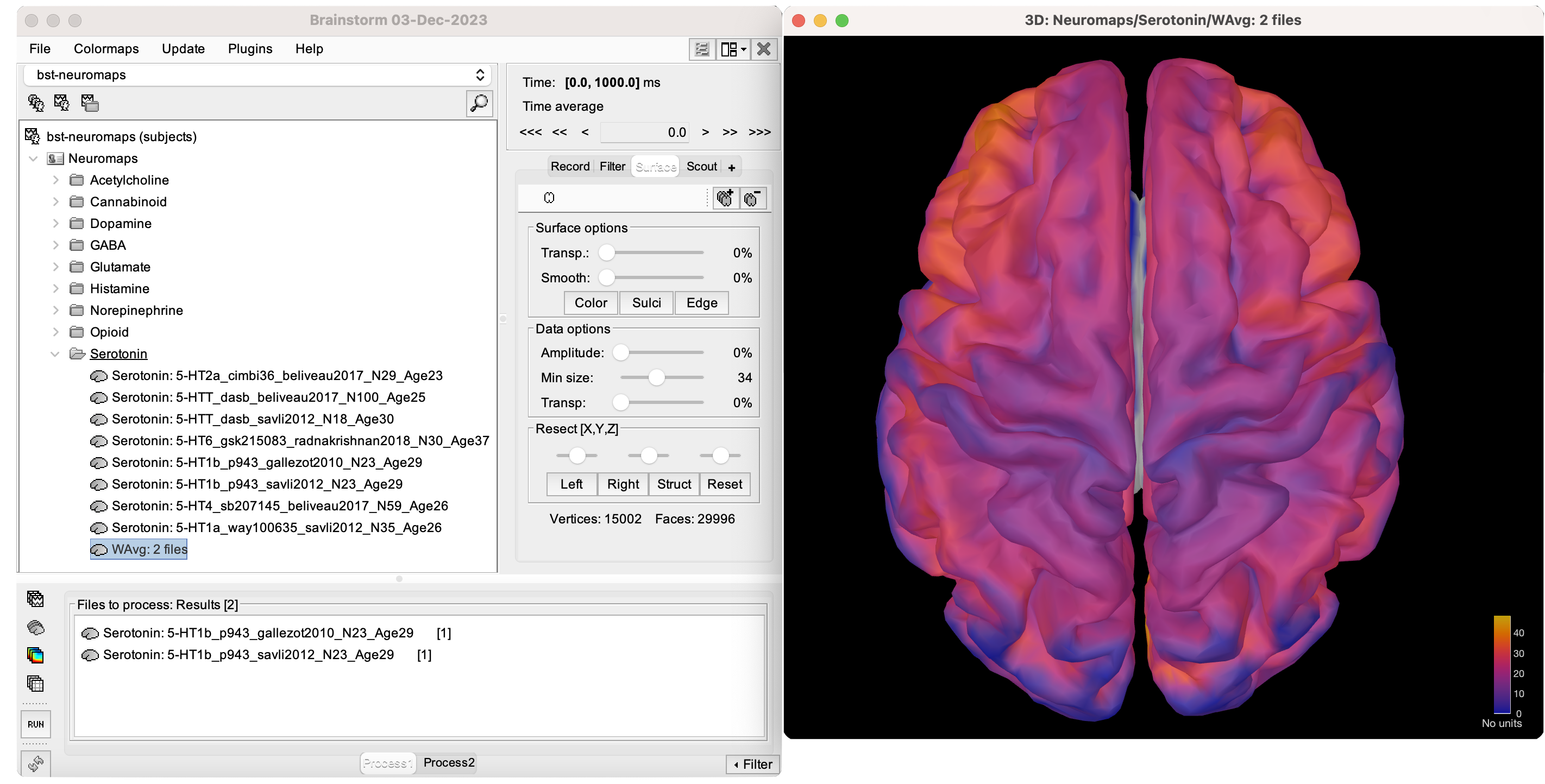
To visualize this new weighted-averaged annotation, follow the same steps as with any brain annotation. Either double-click on the file or right-click > Cortical activations > Display on cortex.
Renaming Files
By default, the new weighted-averaged file is named 'WAvg:' followed by the number of files averaged. If you wish to rename this or any other files from the Neuromaps plugin to provide more informative labels for your analysis, follow these steps:
- Click once on the file to select it.
- With a second click, you will enter renaming mode and the file name will become editable.
- Type in the new desired name for the file.
- Press the "Enter" key on your keyboard or click outside the file name to save the new name.
Alternatively, you can right-click > File > Rename.
Scouting values of annotations
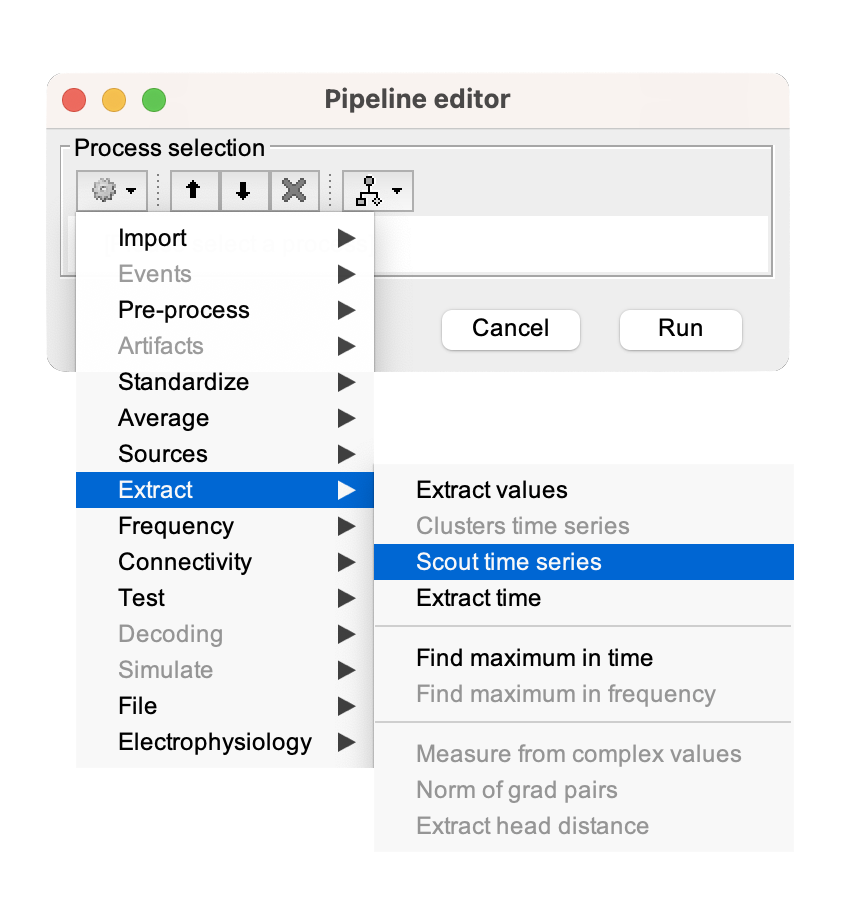
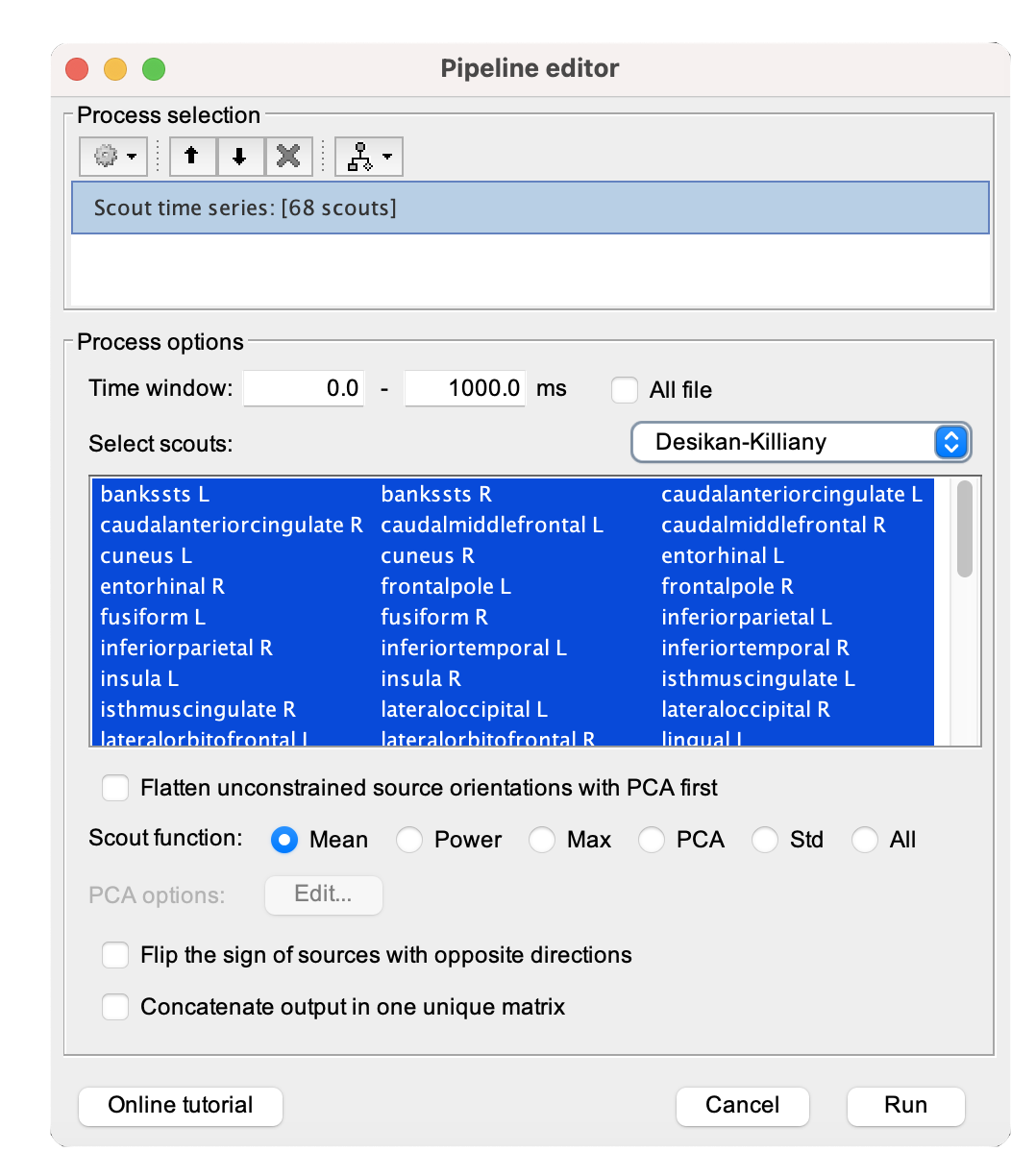
[...] Please refer to Tutorial 23: Scouts for a more in-depth guide in creating, manipulating, and analyzing scouts for effective exploration and comparison of brain activity across different experimental conditions and regions of interest.
Statistical Analyses for Significance Testing
One of the main advantages of having these brain annotations in a common coordinate system is that we can statistically compare their spatial topographies. However, most statistical analyses (e.g., Pearson correlation) assume that the values of observations in each sample are independent of one another. Spatial autocorrelation violates this assumption because samples taken from nearby areas are related to each other and are not independent. Spatially-naive null models—both parametric and non-parametric—yield inflated p-values and are inappropriate for significance testing of neuroimaging brain maps. When applied to spatially-autocorrelated brain maps typical of most neuroimaging data, these models approach a false positive rate of >75% and their usage in the field is discouraged (Markello, R. D., & Misic, B., 2021).
We therefore use spatially informed null models to benchmark the statistical unexpectedness of specific features of interest.
The original neuromaps workflow integrates multiple methods of performing spatial permutations for significance testing. For the plugin, we implemented the most widely adopted of these approaches—the spin method, which involves randomizing the anatomical alignment between two cortical surface maps through spherical rotation by a random angle (Alexander-Bloch et al., 2018).
Spatial correlation with brain annotations
- 1-sample source estimation × brain annotations
- N-sample source estimation × brain annotations
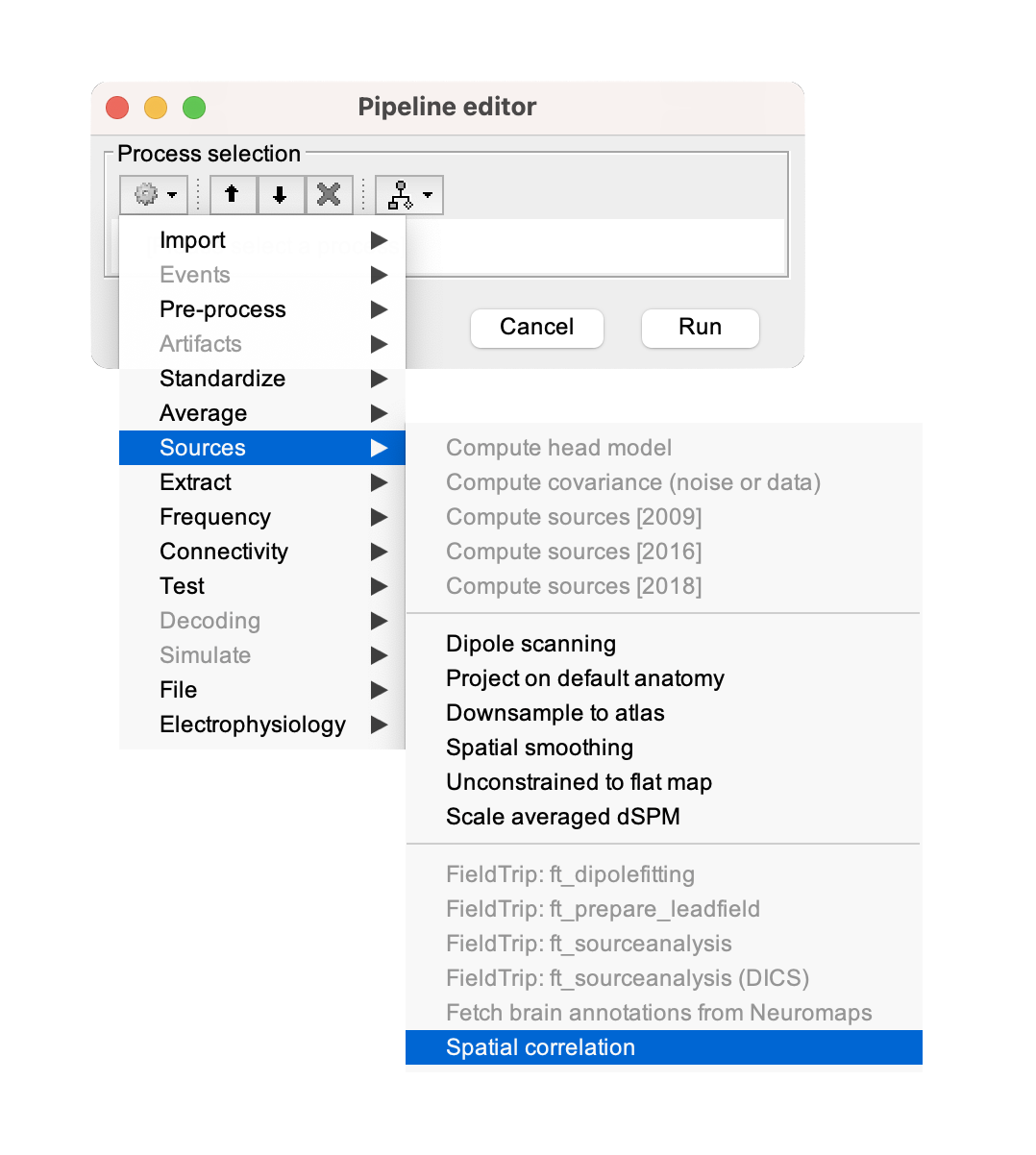
Spatial correlation with any files
- 1-sample source estimation × 1-sample source estimation (or brain annotation)
- N-sample source estimation × 1-sample source estimation (or brain annotation)
- N-sample source estimation × N-sample source estimations
Number of spins
According to the literature, null models converge quickly, reaching stable statistical estimates after approximately 100–500 nulls (Markello, R. D., & Misic, B. 2021). Researchers can use this information to balance accuracy and computational feasibility when determining the number of nulls for their analysis.
Our recommendation is to generate 1000 nulls to establish a reliable null distribution of correlation coefficients and to estimate the two-tailed p-value for the original correlation between the pair of maps.
On the Hard Drive
to be added
Applications of Neuromaps for Integrative Research
The Neuromaps plugin opens up new and exciting avenues for research and can help address questions that depend crucially on anatomical localization. Below, we explore some potential uses of the plugin in advancing integrative research, supported by examples from the literature.
Annotating Structural Connectomes
Hansen, J. Y., Shafiei, G., Voigt, K., Liang, E. X., Cox, S. M., Leyton, M., ... & Misic, B. (2023). Integrating multimodal and multiscale connectivity blueprints of the human cerebral cortex in health and disease. Plos Biology, 21(9), e3002314.
- With the advancements in neuroimaging techniques, which enables the simultaneous exploration of diverse interregional relationships across spatial and temporal scales, neuromaps emerge as an invaluable tool for advancing integrative research in connectomics. The researchers noted a consistent primacy of molecular connectivity modes, demonstrating the mapping of correlated receptor similarity onto multiple phenomena. The use of neuromaps revealed both the similar and complementary ways in which connectivity modes reflect cortical geometry, structure, and disease and assisted in constructing a multimodal wiring map.
Identification of Specific Neurochemical Targets for Potential Future Clinical Treatments
da Silva Castanheira, J., Wiesman, A. I., Hansen, J. Y., Misic, B., Baillet, S., Network Quebec Parkinson, & PREVENT-AD Research Group. (2023). Neurophysiological brain-fingerprints of motor and cognitive decline in Parkinson’s disease. medRxiv.
- Subject-level data can be contextualized against the brain annotations contained in the plugin to produce a subject-specific ‘fingerprint’ of how individuals express different structural and functional brain phenotypes. The authors were able to show the topographical alignment between spectral brain-fingerprints, the functional hierarchy of cortex, and neurotransmitter systems.
Wiesman, A. I., da Silva Castanheira, J., Degroot, C., Fon, E. A., Baillet, S., Network, Q. P., & Prevent-Ad Research Group. (2023). Adverse and compensatory neurophysiological slowing in Parkinson’s disease. Progress in Neurobiology, 231, 102538.
- Neuromaps has the potential to enhance the integration of neurophysiological and neurochemical data for a more comprehensive understanding of brain function, and assist in the identification of specific neurochemical targets for potential future clinical treatments. The authors demonstrated, for instance, that the clinical relationships of neurophysiological slowing in patients with Parkinson's disease are aligned topographically with the densities of selective neurotransmitter systems and the organization of sensory-association areas—namely, dopamine, serotonin, GABA, and norepinephrine systems.
Acknowledgements
All credit for the conception of the original neuromaps algorithm is due to Ross D. Markello, Justine Y. Hansen, Zhen-Qi Liu, Vincent Bazinet, Golia Shafiei, Laura E. Suárez, Nadia Blostein, Jakob Seidlitz, Sylvain Baillet, Theodore D. Satterthwaite, M. Mallar Chakravarty, Armin Raznahan & Bratislav Misic. The appropriate citation for neuromaps is as follows:
Markello, R.D., Hansen, J.Y., Liu, ZQ. et al. neuromaps: structural and functional interpretation of brain maps. Nat Methods 19, 1472–1479 (2022).
If you used any of the included maps, please also cite the original papers that publish the data. References for each map can be found in this spreadsheet.
- If you used the surface maps which were transformed using the registration fusion framework, please also cite:
Wu, J. et al. Accurate nonlinear mapping between MNI volumetric and FreeSurfer surface coordinate systems. Hum. Brain Mapp. 39, 3793–3808 (2018).
Contributing
We welcome contributions from the community to help improve and expand the functionality of the Neuromaps plugin. Feel free to submit pull requests on the Github repository, report issues, or provide suggestions below! Your feedback is invaluable in ensuring a user-friendly experience for researchers worldwide. We believe that open science is most impactful when the countless everyone is provided with equal access to the newest and greatest resources in the field.