PET processing in Brainstorm
Authors: Diellor Basha
Contents
Introduction
This tutorial describes how to process and analyze Positron Emission Tomography (PET) data within Brainstorm and how to extend PET data to surface-based, multimodal analysis. It guides users through importing and pre-processing PET volumes and outlines steps for surface-based analysis of cortical tracer uptake values. PET is a powerful imaging technique widely used in medical and scientific research to study metabolic and functional processes in the brain and body. PET processing involves a series of computational and analytical steps to transform raw data into meaningful visualizations and quantitative insights. These steps ensure accurate reconstruction, correction, and analysis of PET scans, enabling researchers and clinicians to draw precise conclusions. The increasing availability of multimodal datasets that combine PET, MRI and M/EEG provide unique opportunities for integrated analysis of neurophysiology, brain structure and molecular processes mapped by PET radioligands.
This page provides an overview of the essential methods, tools, and considerations involved in PET processing, aiming to support both newcomers and experienced users of Brainstorm. If you are new to Brainstorm, refer to the comprehensive introductroy materials in Brainstorm introduction tutorials to familiarize yourself with the layout and workflow in Brainstorm.
Method Overview
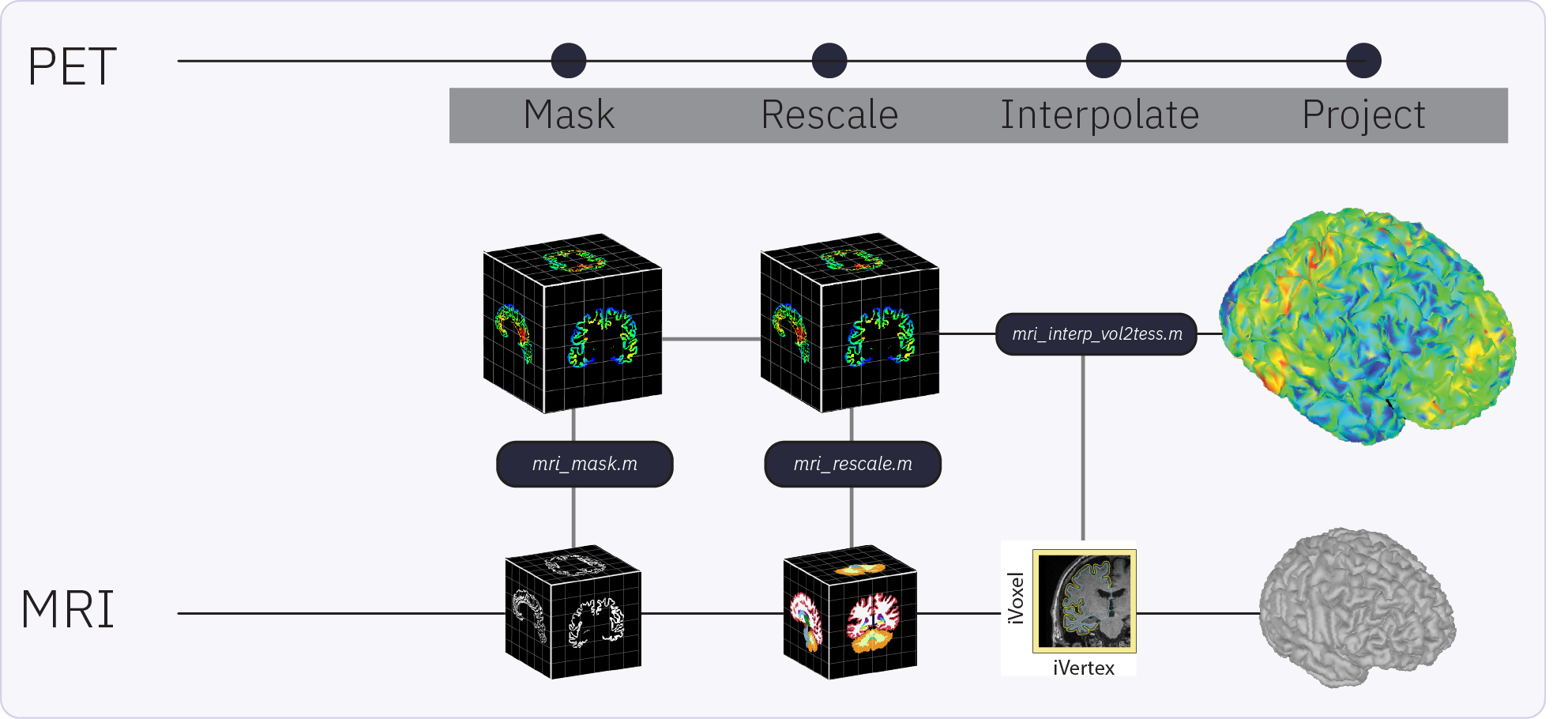
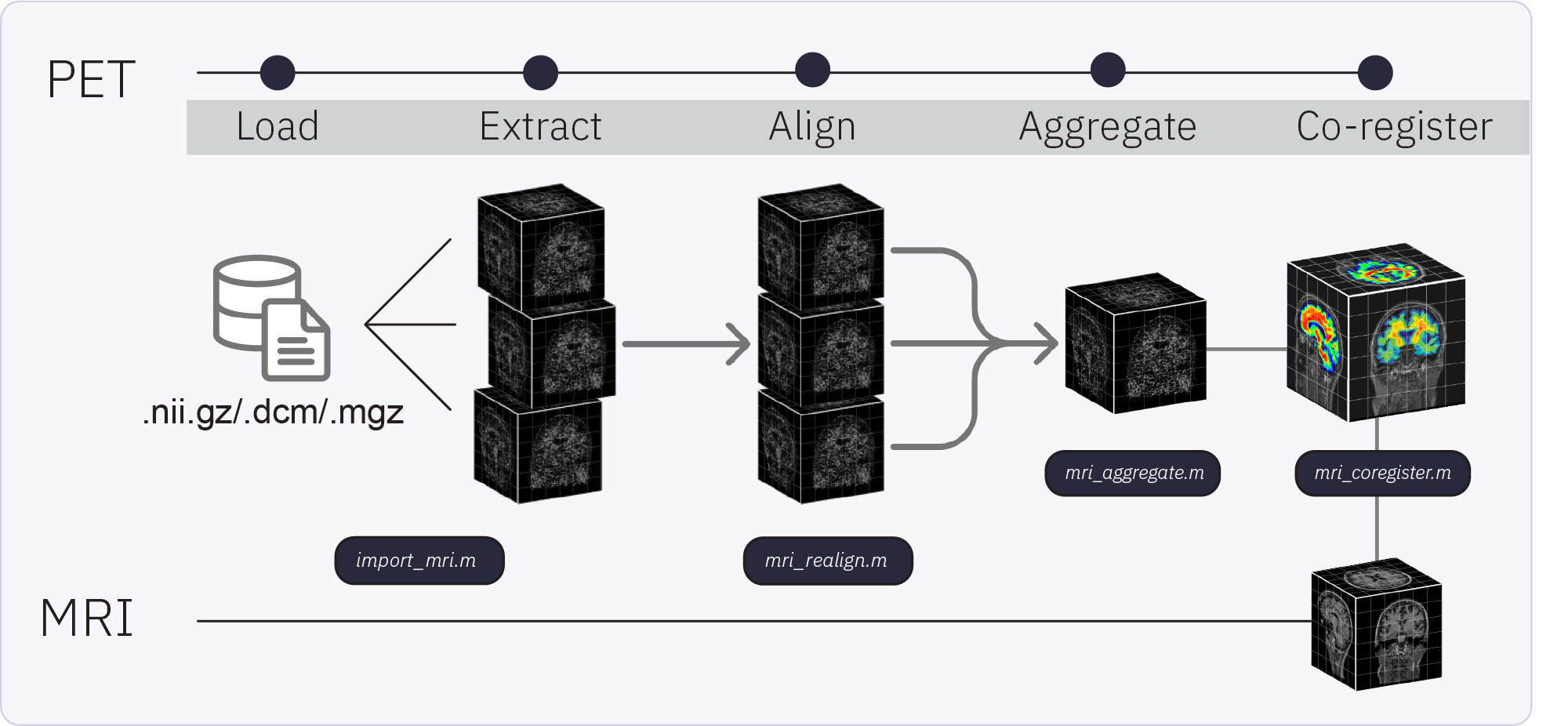
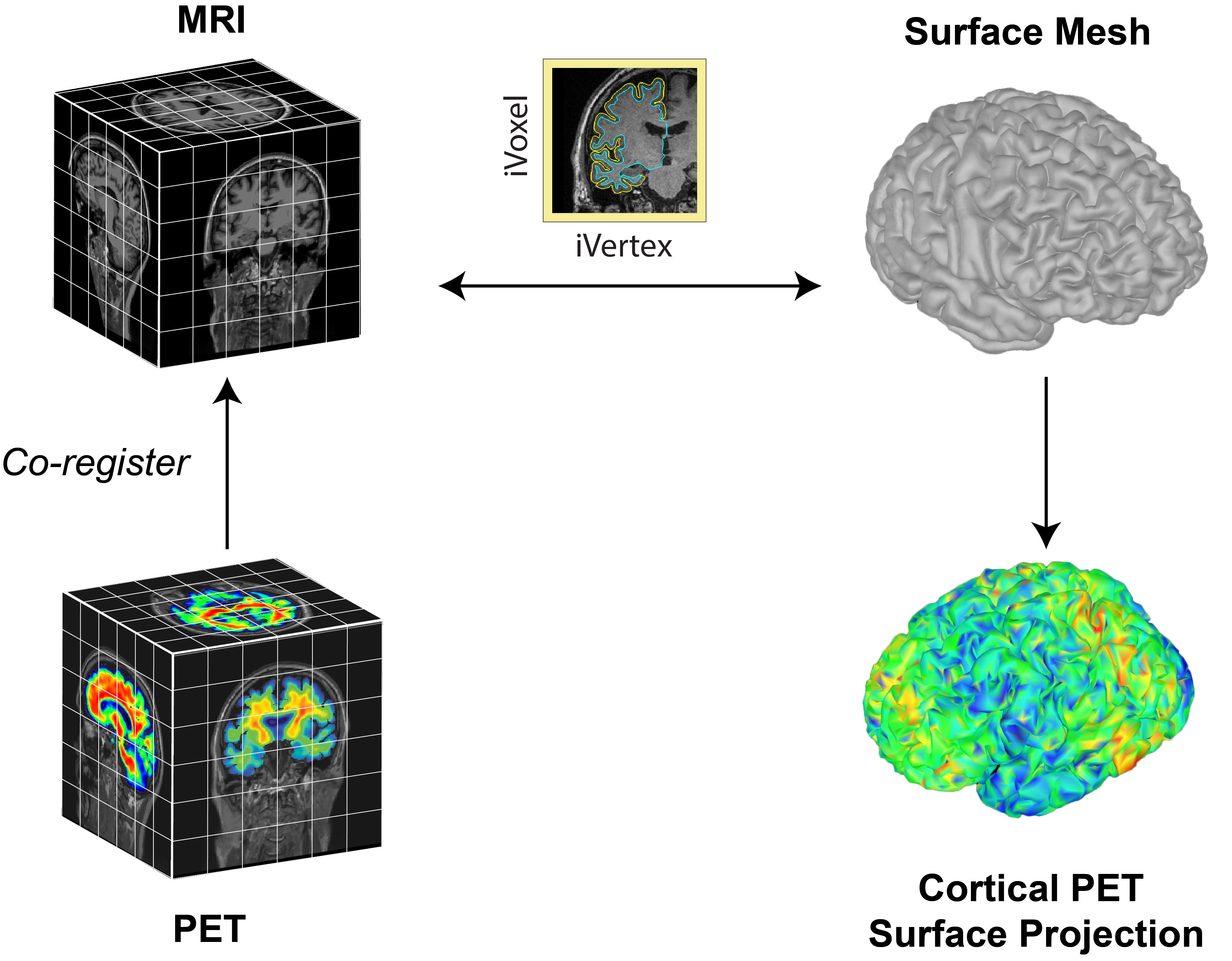
The Brainstorm extension for PET supports importing, processing, registering, visualizing, and analyzing PET data within Brainstorm. PET functionality and workflow are designed to facilitate the analysis of multimodal neuroimaging data by allowing the user to co-analyze MEG, PET and MRI-derived data. Multi-frame (4D) PET volumes are realigned during import and aggregated over time from the 4D image (i.e. mean of the n frames) to obtai a unique 3D volume for co-registration with the subject’s structural MRI. Multi-frame PET volumes are realigned using SPM’s realign and reslice functions and co-registered with SPM. The aggregated and co-registered 3D volume is masked and rescaled to obtain voxel-wise standardized uptake value ratio (SUVR) which are then projected to the cortical surface.
Dataset and requirements
Prerequisites:
Brainstorm Installation: Ensure you have a working copy of Brainstorm installed on your computer.
SPM12 (Statistical Parametric Mapping): required for realignment of dynamic (4D) PET volumes and co-registration of PET with MRI.
Files in dataset
tutorial_pet_processing/
mri/: Anatomy data
Subject01_T1.nii.gz: Raw T1 MRI of subject 1 (in NIfTI-1 format),
freesurfer/: Folder containing reconstructions of the cortical surface and tissue segmentations, precomputed from the MRI volumes above using Freesurfer. See * Using Freesurfer for more details about computing Freesurfer reconstructions from Brainstorm.
pet/: PET data
Subject01_trc-18FNAV4694_pet.nii.gz: Raw PET data taken over 6 acquisition frames, with uptake values for beta-amyloid tracer 18FNAV4694
Subject01_trc-18Fflortaucipir_pet.nii.gz: Raw PET data taken over 4 acquisition frames, with uptake values for tau tracer 18Fflortaucipir
Import the anatomy
- Start Brainstorm
Select the menu File > Create new protocol. Name it "pet_processing" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
Reference MRI
Go to the Anatomy view
Right-click on the pet_processing top node > New subject > Subject01.
Keep the default options you defined for the protocol.Switch to the Anatomy view of the protocol.
Right-click on the subject node > Import MRI.
Set the file format: MRI: NIfTI-1 (*.nii;*.nii.gz)???.
Select: Subject01_T1.nii.gzThe MRI viewer opens automatically. Click on "Click here to compute MNI normalization", option "maff8".
Click on Save to close the MRI viewer. New node named Subject01_T1 is created.
Import and pre-process the PET volume
The MRI volume above will be used as the anatomical reference for this subject. We will now import two PET scans done on the same subject and process the raw PET data.
Right-click on the subject node > Import PET.
Select: Subject01_trc-18FNAV4694_pet.nii.gzChoose Yes for the transformation for MRI orientation.
- Choose the import options for PET
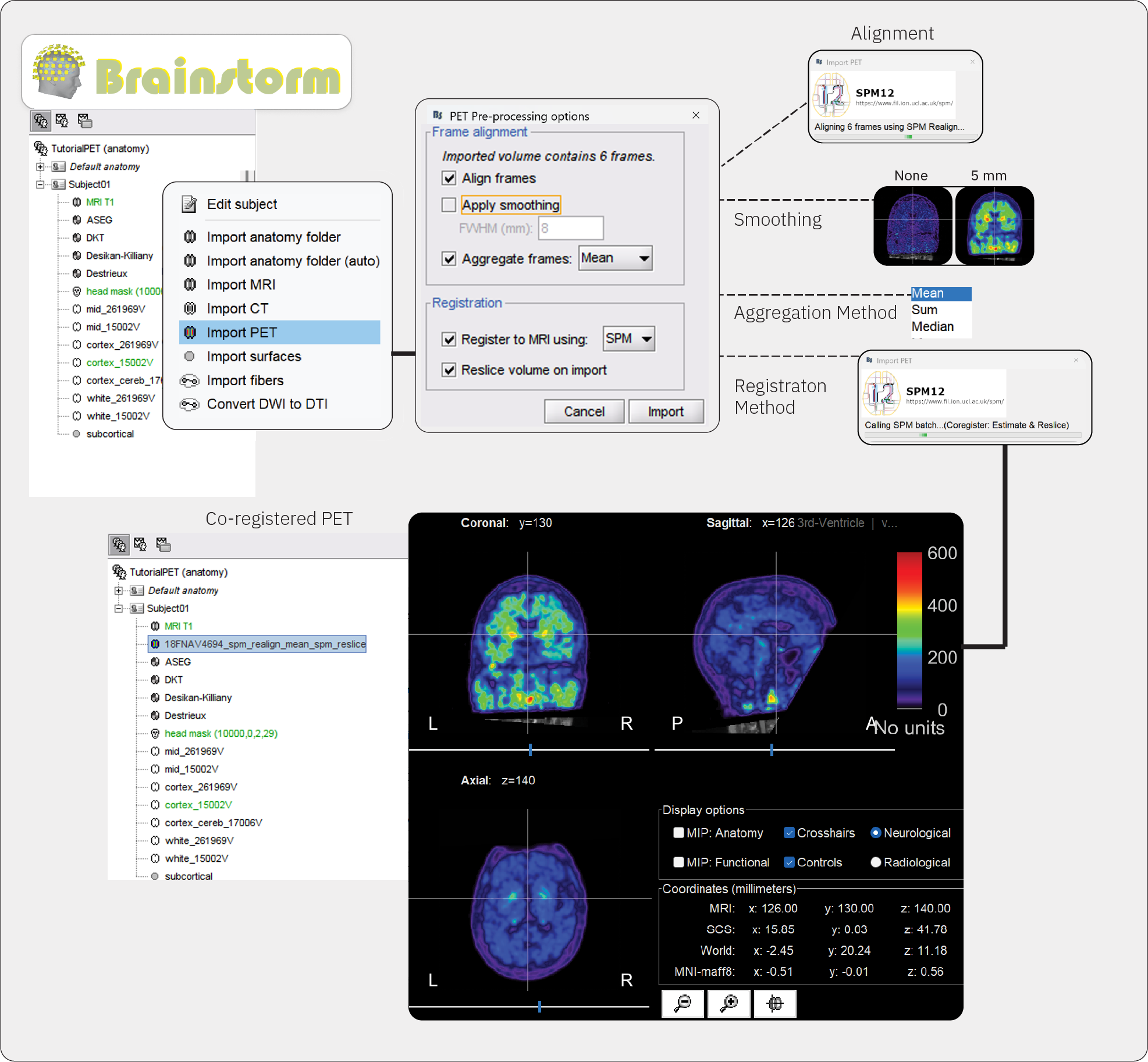
Align frames: If the imported file is a multi-frame (dynamic) volume, realignment is recommended before aggregation and registration to MRI. Brainstorm checks automatically whether the imported file is a 3D (single-frame) or 4D (multi-frame) scan and makes available the option for realignment accordingly (greyed out if single-frame). In this step, individual PET frames are first aligned to a chosen target frame (the first) and then realigned in a second pass to the mean of all frames available in the volume. In this case, Subject01_trc-18FNAV_pet.nii.gz is a 4D (dynamic) structure, containing a PET scan with 6 acquisition frames, taken minutes apart.
Check this optionApply smoothing: This option applies Gaussian smoothing to uptake values in each PET frame before frames are aggregated into a single 3D volume. The size of the smoothing parameter is the full width at half maximum (FHWM) of the smoothing function, given in mm. Smoothing at this stage can exarcebate partial fraction effects, whereby non-specific tracer binding from white matter and low uptake in cerebrospinal fluid (CSF) can contaminate the cortical signal. In some cases with low-resolution PET scans, smoothing at this step may be necessary to improve registration to MRI. We recommend skipping smoothing at this stage and smoothing instead on the surface at later steps during surface-based processing described below.
Uncheck this option or set FWHM to 0 mm.Aggregate frames: This option lets you choose how the realigned frames will be aggregated to produce the time-aggregated, single-frame 3D volume.
Select meanRegister to MRI using: The imported PET volume needs to be registered to the reference MRI in order to align tracer uptake values to the corresponding anatomy. It is a necessary step for both volume and surface-based analysis of tracer uptake. Refer to the CT2MRI tutorial for detailed explanation of the two co-registration methods: SPM and MNI.
Check this option, and set method to SPMReslice volume: This option resamples the PET volume to match the slice sampling of the reference MRI. It is a necessary step for overlaying PET slices to their on the reference MRI. If you know that your imported PET volume is already co-registered and resliced to the reference MRI, select No for this option. For the current dataset, we will reslice the volume.
Check this option
You can also decide to not perform any of these actions, and perform them once the PET volume is imported using the PET volume context menu:

Once processing and importing is completed, the MRI viewer will open automatically, displaying the PET volume as an overlay over the reference MRI. The default colormap of the PET overlay can be modified by right-clicking on the figure and selecting from the popup menu. Use the viewer to explore the PET data and to verify that co-registration and pre-processing was done accurately.
Repeat the above steps for the second PET file Subject01_trc-18Fflortaucipir_pet.nii.gz.
Masking and rescaling PET
The imported PET volume will now appear in the Brainstorm tree view under its own PET icon with PET-specific context menus. To compute the standardized uptake value ratio (SUVR), we will rescale PET uptake values with respect to a reference region such as the cerebellum. As this is a ROI-based method, accurate parcellation of the volume is required before proceeding. You can compute tissue segmenation and parcellation from the MRI file within Brainstorm. For detailed tutorials, refer to MRI segmentation in https://neuroimage.usc.edu/brainstorm/Tutorials/#Advanced_tutorials
For the purposes of this tutorial, we will skip the long segmentation step and use the precomputed Freesurfer segmentation available in the dataset. Select Subject01 > Import anatomy folder (auto) , choose Freesurfer + Volume atlases + Thickness from the file type menu and select the precomputed Freesurfer folder above.
We will use the precomputed Freesurfer segmentation and parcellation to obtain the average uptake value in a selected ROI.
Go to the Anatomy view
Right-click on the Subject01_trc-18Fflortaucipir_pet node > PET processing > Compute SUVR.
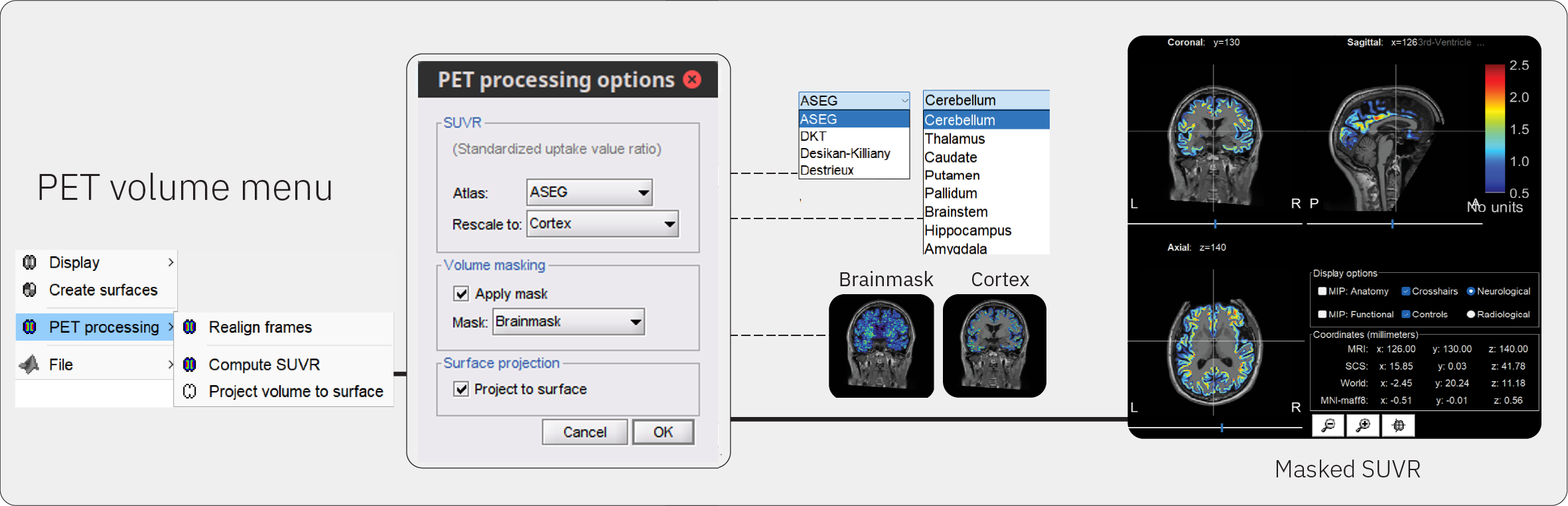
Reference ROI: This is the brain region that will be used as a reference for rescaling PET values to obtain the SUVR. Select Cerebellum from the drop down menu.
Atlas: Brainstorm stores multiple atlas files that contain different methods of parcellation. Select ASEG to use the Freesurfer parcellation.
Apply mask: The rescaled PET data can be masked to restrict PET values to a chosen ROI mask. Select Cortex from the drop down menu.
Project to surface: This option allows you to project cortical PET values to the cortical surface for surface-based analysis.
Surface-based analysis
The surface-based submodule enables the projection of volumetric PET data onto the cortical surface, facilitating vertex-wise analysis in a common subject framework. Surface-based analysis of PET is based on the tess2mri interpolation matrix computed by Brainstorm. This matrix establishes weights for volumetric MRI voxels contributing to specific vertices on the cortical surface. Weights range between 0 (no contribution) and 1 (full contribution), enabling a smooth mapping of voxel coordinates to the cortical surface. Given that PET data are co-registered to the MRI, PET voxel intensity values are scaled by the interpolation weights, resulting in a weighted projection of PET values to the nearest vertex.
Partial Volume Correction
Related tutorials
Articles
Forum discussions
Overlaying PET on cortex: https://neuroimage.usc.edu/forums/t/30241