T1-MRI Segmentation with SPM12 / CAT12
Authors: Francois Tadel
 CAT is a SPM12 toolbox that is fully interfaced with Brainstorm. It can replace efficiently FreeSurfer for generating the cortical surface from any T1 MRI. It runs on any OS in about 1 hour, instead of the typical 24hr FreeSurfer recon-all processing. The surfaces are registered to the templates with the FreeSurfer spheres, and include many anatomical and functional atlases.
CAT is a SPM12 toolbox that is fully interfaced with Brainstorm. It can replace efficiently FreeSurfer for generating the cortical surface from any T1 MRI. It runs on any OS in about 1 hour, instead of the typical 24hr FreeSurfer recon-all processing. The surfaces are registered to the templates with the FreeSurfer spheres, and include many anatomical and functional atlases.
You can either run the CAT segmentation from Brainstorm, or run it separately and import its outputs as you would do with FreeSurfer. The software is not packaged with Brainstorm and does not work with the compiled version of Brainstorm, you must install SPM12 and CAT separately.
Contents
Install SPM12
Download SPM12: https://www.fil.ion.ucl.ac.uk/spm/software/download/
Unzip it anywhere on your computer, but not in any of the Brainstorm folder.
- Do not add the spm12 folder to your path if you are not a SPM user, Brainstorm will do this automatically when needed.
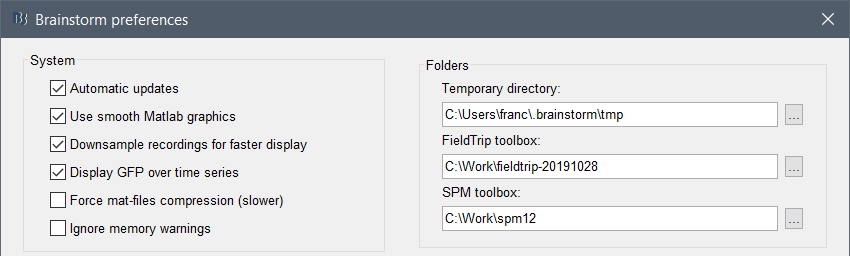
Start Brainstorm, set the SPM12 path in the Brainstorm preferences (File > Edit preferences).
Install CAT12
Download CAT: http://www.neuro.uni-jena.de/cat/index.html#DOWNLOAD
- Unzip the downloaded file.
Move the cat12 folder to the spm12/toolbox directory.
Citing CAT12
If you use CAT from Brainstorm for MRI segmentation, please cite the following article in your publications:
Gaser C, Dahnke R, Kurth F, Luders E
CAT - A Computational Anatomy Toolbox for the Analysis of Structural MRI Data
NeuroImage, in review
Running CAT from Brainstorm
- Switch to the anatomy side of the database explorer.
- Create a subject, import the T1 MRI for this subject.
Set the fiducial points manually (NAS/LPA/RPA) or compute the MNI transformation.
Right-click on the MRI > CAT12 MRI registration,
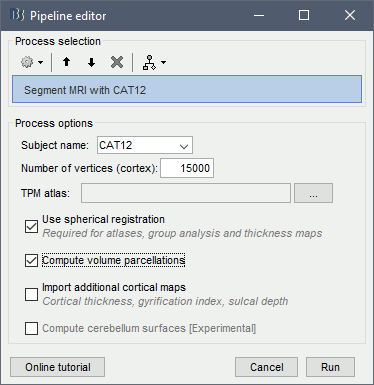
or use the process Import anatomy > Segment MRI with SPM12/CAT12 (more options)
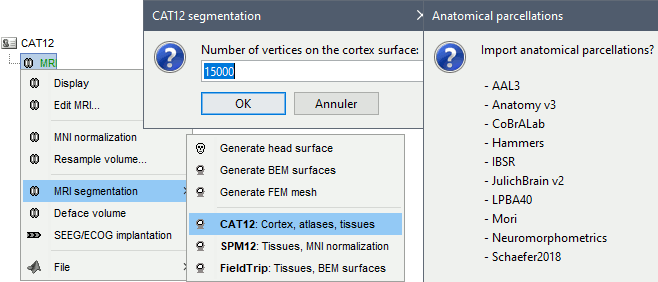
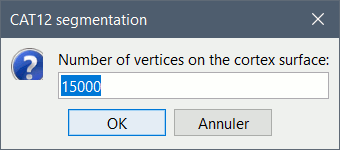
In interactive mode, you will be prompted for the number of vertices you want in the final cortex surface. This will by extension define the number of dipoles to estimate during the source estimation process. By default we set this value to 15000 for the entire brain (it means 7500 for each hemisphere).
- With the process version, you have access to more options:
TPM atlas: Location of the template tissue probabilistic maps. If empty, it uses by default the standard TPM.nii available in SPM12 (usually downloaded automatically in the folder $HOME/.brainstorm/defaults/spm/TPM.nii). It can be interesting to replace it with a probabilistic atlas better adapted to specific population, eg. the children atlas from CAT12 (spm12/toolbox/cat12/templates_volumes/TPM_Age11.5.nii)
Use spherical registration: Call CAT12 with the highest possible accuracy, which includes the spherical registration to the FSAverage template. Enabling this option takes much longer, but is necessary for importing all the FreeSurfer atlases, projecting the sources maps to a common template in the case of group analysis, and computing accurate cortical thickness maps.
Import cortical thickness maps: Enable this option to import the cortical thickness computed by CAT12 as source files. See section Cortical thickness.
Brainstorm saves the MRI to process in a temporary folder, then calls CAT as an SPM batch:
$HOME/.brainstorm/tmp/cat12/spm_cat12.nii- All the output from CAT is saved in the same temporary folder. At the end, Brainstorm imports the output folder as the anatomy for the selected subject. If the process crashes, you can inspect the contents of this folder for indices on how to solve the problem. If the segmentation and the import is successful, the temporary folder is deleted.
If you get an error about a missing function spm_ov_mesh.m, you need to update SPM12 first (run "spm_update" from the Matlab command line), or download the missing file (https://github.com/spm/spm12/blob/master/spm_orthviews/spm_ov_mesh.m) from Github in folder spm12/spm_orthviews.
https://neuroimage.usc.edu/forums/t/cat12-segmentation-error-in-linux-and-windows/13409/2
A report is displayed by CAT, and saved as an image and a PDF file in tmp/cat12/report.
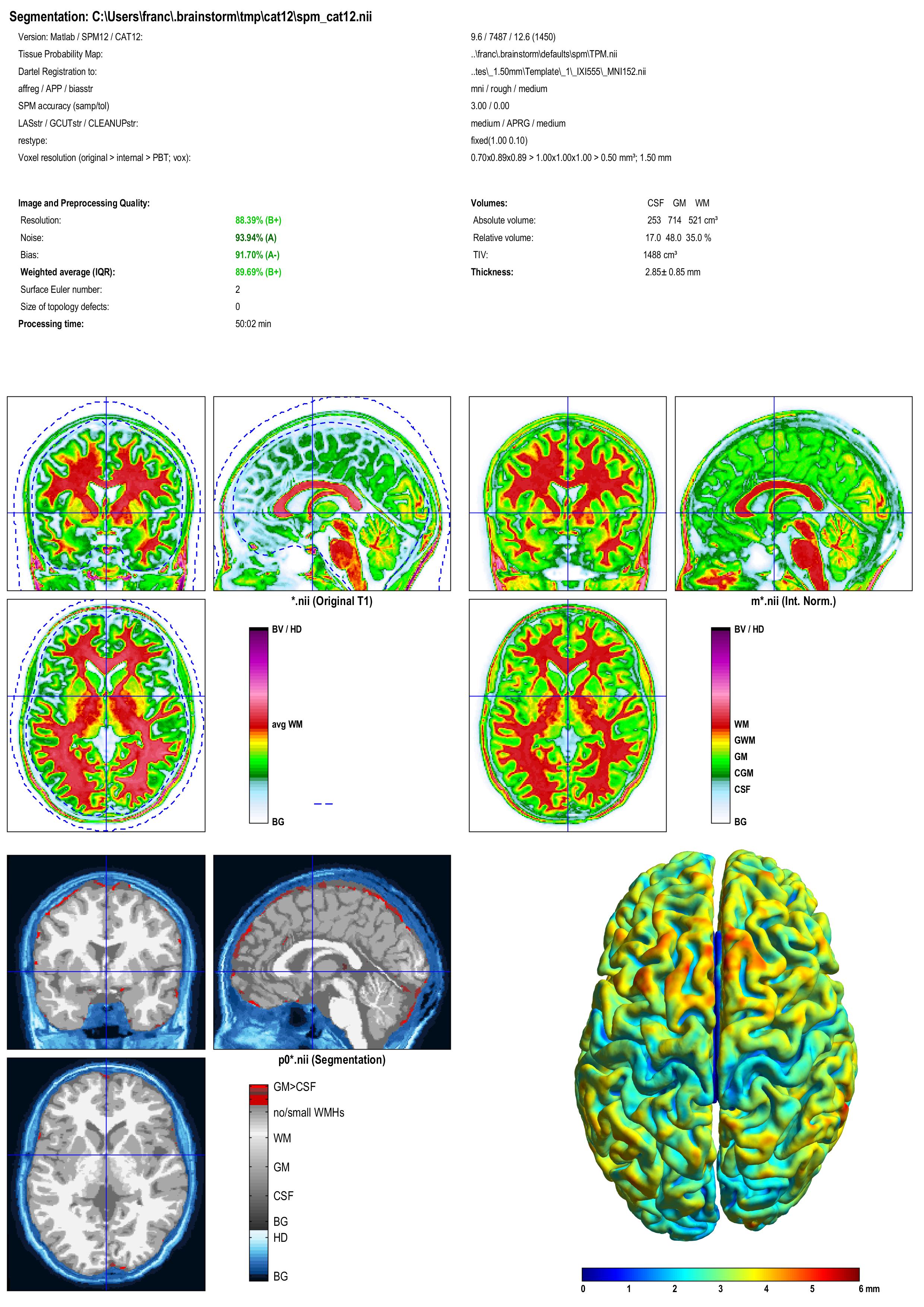
Importing the results in Brainstorm
If you run the segmentation process from Brainstorm, the import will be done automatically. Otherwise, if you need to import an existing CAT segmentation, here is the following procedure.
Right-click on the subject > Import anatomy folder.
Select the file format CAT12 folder or CAT12 folder + thickness maps.
- Enter the number of vertices in the final cortex surface.
- The files that are imported from the segmentation output folder are the following:
/*.nii (T1 MRI volume - only one .nii file allowed in the top-level folder)
/surf/?h.central.*.gii (left/right hemisphere of the central surface)
/surf/?h.sphere.reg.*.gii (left/right FreeSurfer registered spheres)
/surf/?h.thickness (texture with the cortical thickness at each vertex)
Additional atlases are imported from the CAT12 program folder (spm12/toolbox/cat12):
/templates_surfaces/?h.central.freesurfer.gii (FSAverage central surfaces, high resolution)
/templates_surfaces_32k/?h.central.freesurfer.gii (FSAverage surfaces, medium resolution)
/atlases_surfaces/?h.*.annot (cortical surface-based atlases)
/atlases_surfaces_32k/?h.*.annot (cortical surface-based atlases)
All these atlases available on the FSAverage template are projected to the subject native surfaces using the FreeSurfer registered spheres, with functions bst_project_scouts.m and tess_interp_tess2tess.m.
The central surfaces generated by CAT are meshes half-way between the grey-white interface and the external pial surface.
- The files you can see in the database explorer at the end:
MRI: The T1 MRI of the subject, imported from the .nii file at the top-level folder.
head mask (10000,0,2): Scalp surface generated by Brainstorm. The numbers indicate the parameters that were automatically used for this head: vertices=10000, erode factor=0, fill holes=2.
cortex_250000V: High-resolution cortex surface that was generated by CAT.
cortex_15000V: Low-resolution cortex surface, downsampled using the reducepatch function from Matlab (it keeps a meaningful subset of vertices from the original surface). It appears in green in the database explorer, ie. it is going to be used as the default by the processes that require a cortex surface.
A figure is automatically shown at the end of the process, to check visually that the cortex and head surfaces were properly generated and imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.
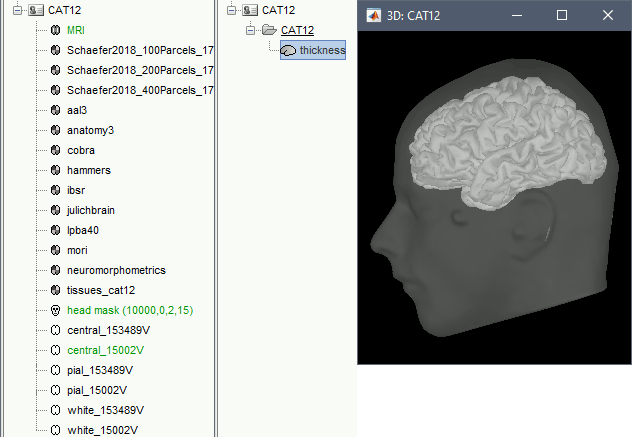
Cortical parcellations
FreeSurfer's FSAverage subject includes parcellations of the cortical surface in anatomical or functional regions. The description of this feature is available here:
http://freesurfer.net/fswiki/CorticalParcellation
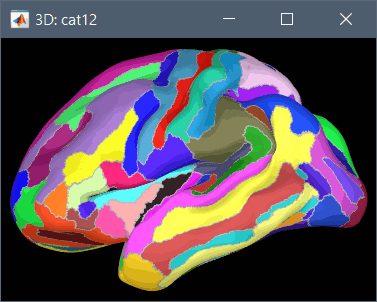
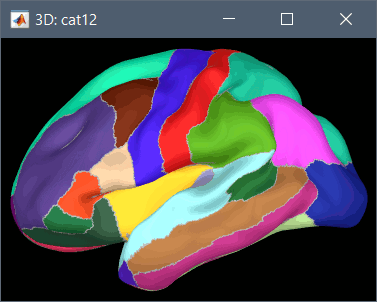
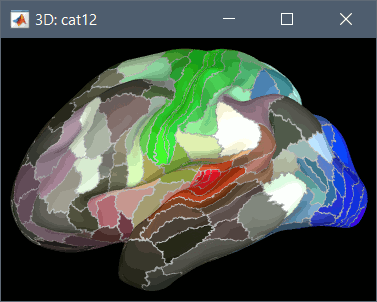
Using CAT12 from Brainstorm, you have access to 15 atlases on all the individual brains:
Destrieux atlas (atlases_surfaces/?h.aparc_a2009s.freesurfer.annot): more info
Desikan-Killiany atlas (atlases_surfaces/?h.aparc_DK40.freesurfer.annot): more info
HCP MMP1 atlas (atlases_surfaces/?h.aparc_HCP_MMP1.freesurfer.annot): more info
Schaefer 2018 atlases (atlases_surfaces_32k/?h.Schaefer2018_*.annot): more info
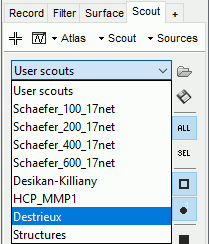
These atlases are imported in Brainstorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution cortex file > File > View file contents. You can see that 17 structures "Atlas" are available, the first one that has Name='User scouts', and the second one Name='Destrieux'.
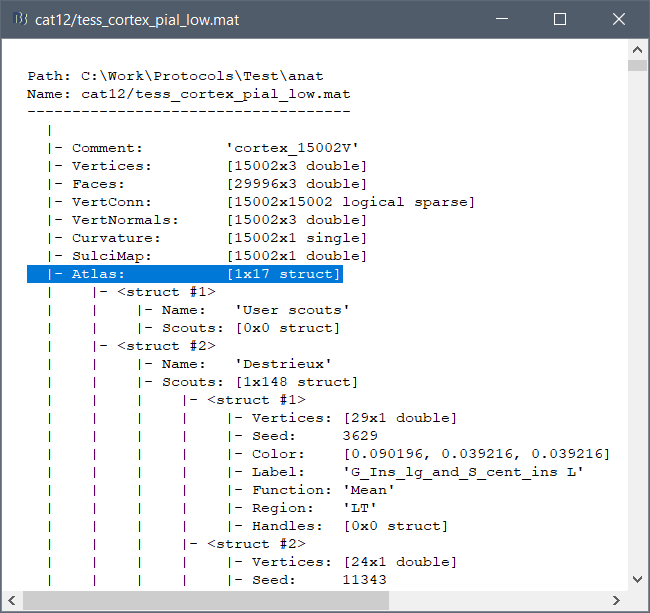
To access them from the interface: Double-click on the cortex and go to the Scout tab, and click on the drop-down list to select another Atlas (ie group of scouts):
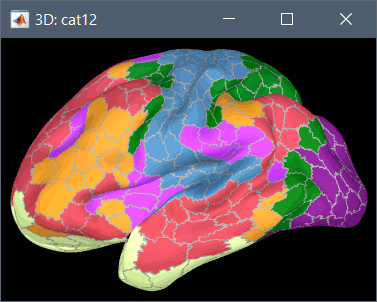
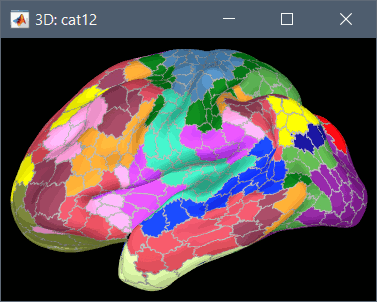
Cortical thickness
The cortical thickness can be saved as a cortical map in the database (a "results" file). This result is generated when using the file format "CAT12 folder + Thickness maps" in the Import anatomy folder selection.
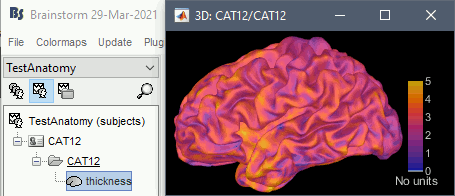
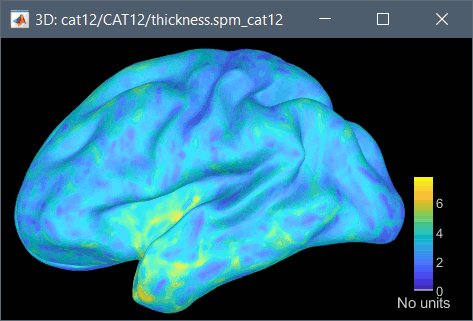
SPM batch
The CAT segmentation is executed with the following SPM12 batch:
matlabbatch{1}.spm.tools.cat.estwrite.data = {[NiiFile ',1']};
matlabbatch{1}.spm.tools.cat.estwrite.data_wmh = {''};
matlabbatch{1}.spm.tools.cat.estwrite.useprior = '';
matlabbatch{1}.spm.tools.cat.estwrite.nproc = 0; % Blocking call to CAT12
matlabbatch{1}.spm.tools.cat.estwrite.opts.tpm = {TpmNii}; % User-defined TPM atlas
matlabbatch{1}.spm.tools.cat.estwrite.output.bias.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.GM.native = 1; % GM tissue maps
matlabbatch{1}.spm.tools.cat.estwrite.output.GM.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.GM.mod = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.GM.dartel = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.WM.native = 1; % WM tissue maps
matlabbatch{1}.spm.tools.cat.estwrite.output.WM.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.WM.mod = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.WM.dartel = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.TPMC.native = 1; % Tissue classes 4-6 to create own TPMs
matlabbatch{1}.spm.tools.cat.estwrite.output.TPMC.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.TPMC.mod = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.TPMC.dartel = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.CSF.native = 1; % CSF tissue maps
matlabbatch{1}.spm.tools.cat.estwrite.output.CSF.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.CSF.mod = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.CSF.dartel = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.label.native = 1; % Label: background=0, CSF=1, GM=2, WM=3, WMH=4
matlabbatch{1}.spm.tools.cat.estwrite.output.label.warped = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.label.dartel = 0;
% Volume atlases
if isVolumeAtlases
matlabbatch{1}.spm.tools.cat.estwrite.output.warps = [1 1]; % Save non-linear MNI normalization deformation fields: [forward inverse]
matlabbatch{1}.spm.tools.cat.estwrite.output.ROI = 1;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.neuromorphometrics = 1;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.lpba40 = 1;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.cobra = 1;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.hammers = 1;
matlabbatch{1}.spm.tools.cat.estwrite.output.atlas.native = 1; % Save atlases in native space
% Add all other MNI atlases available in Brainstorm
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.ownatlas = {...
'C:\Work\RawData\Test\segment\Atlases\cat_aseg\aparc_a2009s.nii,1'; ...
'C:\Work\RawData\Test\segment\Atlases\cat_aseg\aparc_DKT40.nii,1'; ...
};
else
% Skip deformation fields for the moment: [forward inverse]
matlabbatch{1}.spm.tools.cat.estwrite.output.warps = [0 0];
% No ROIs
matlabbatch{1}.spm.tools.cat.estwrite.output.ROI = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.noROI = struct([]);
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.neuromorphometrics = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.lpba40 = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.cobra = 0;
matlabbatch{1}.spm.tools.cat.estwrite.output.ROImenu.atlases.hammers = 0;
end
% Spherical registration (much slower)
if isSphReg && ~isCerebellum % CGaser comment: Cerebellum extraction is experimental, not to be used routinely
matlabbatch{1}.spm.tools.cat.estwrite.output.surface = 1; % 1: lh+rh
elseif isSphReg && isCerebellum
matlabbatch{1}.spm.tools.cat.estwrite.output.surface = 2; % 2: lh+rh+cerebellum
elseif ~isSphReg && ~isCerebellum
matlabbatch{1}.spm.tools.cat.estwrite.output.surface = 5; % 5: lh+rh (fast, no registration, quick review only)
elseif ~isSphReg && isCerebellum
matlabbatch{1}.spm.tools.cat.estwrite.output.surface = 6; % 6: lh+rh+cerebellum (fast, no registration, quick review only)
end
% Extract additional surface parameters: Cortical thickness, Gyrification index, Sulcal depth
if isExtraMaps
% Separate SPM process (second element in the batch)
matlabbatch{2}.spm.tools.cat.stools.surfextract.data_surf(1) = cfg_dep('CAT12: Segmentation (current release): Left Central Surface', substruct('.','val', '{}',{1}, '.','val', '{}',{1}, '.','val', '{}',{1}, '.','val', '{}',{1}), substruct('()',{1}, '.','lhcentral', '()',{':'}));
matlabbatch{2}.spm.tools.cat.stools.surfextract.GI = 1; % Gyrification index
matlabbatch{2}.spm.tools.cat.stools.surfextract.SD = 1; % Sulcal depth
matlabbatch{2}.spm.tools.cat.stools.surfextract.nproc = 0; % Blocking call to CAT12
end
% Run SPM batch
spm_jobman('initcfg');
spm_jobman('run',matlabbatch);
Additional documentation
Git issue: Bugs CAT12 v1728