Elekta-Neuromag tutorial: Median nerve stimulation
This tutorial describes how to process continuous Elekta-Neuromag MEG recordings. It is based on median nerve stimulation acquired at the Massachusetts General Hospital in 2005 with a Neuromag Vectorview 306 system. The sample dataset contains the results for one subject for both arms: 18 minutes, 300 stimulations per arm.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Contents
License
This data was collected in a multi-site MEG study administered by the MIND institute.
Presentation of the experiment
Experiment
- One subject, one acquisition run of 18 minutes
- The run contains 624 electric stimulations randomly distributed between left and right:
- 301 stimulations of the left hand
- 323 stimulations of the right hand
MEG acquisition
Acquisition at 1793Hz, with a Neuromag Vectorview 306 system
- Recorded at the Massachusetts General Hospital in 2005
- Recorded channels (318):
- 102 MEG magnetometer
- 204 MEG planar gradiometers
- 9 stimulation channels (#307-315)
- 2 EOG bipolar (#316-317)
- 1 ECG bipolar (#318)
1 dataset: mind004_050924_median01_raw.fif
Head shape
- 130 additional head points
Subject anatomy
- Subject with 1.5T MRI
Processed with FreeSurfer 5.2
Download and installation
Requirements: You have already followed all the basic tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website and download the file sample_neuromag.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version)
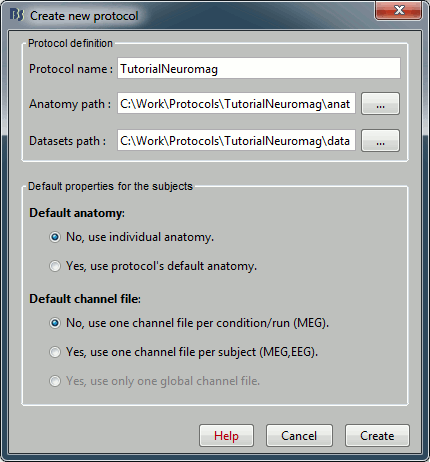
Menu File > Create new protocol > TutorialNeuromag, with the following options:
No, use individual anatomy,
No, use one channel file per run.
Import the anatomy
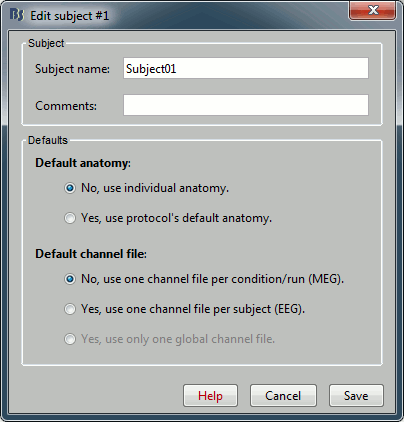
Right-click on the TutorialNeuromag folder > New subject > Subject01 (leave all the defaults).
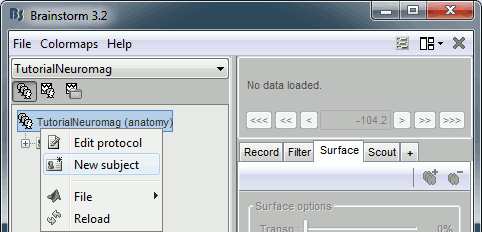
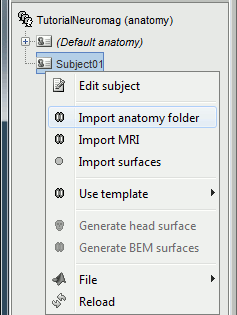
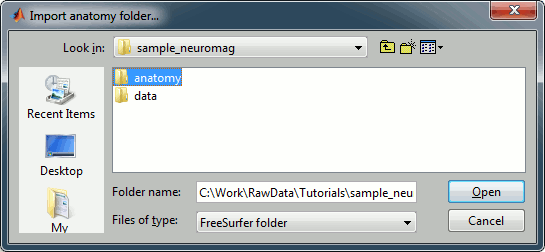
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: sample_neuromag/anatomy
Number of vertices of the cortex surface: 15000 (default value)
Click on the link "Click here to compute MNI normalization".
- Set the 3 required fiducial points (indicated in MRI coordinates):
- NAS: x=131, y=232, z=123
- LPA: x=48, y=136, z=74
- RPA: x=204, y=131, z=67
At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.
Access the recordings
Link the recordings
- Switch to the "functional data" view, the middle button in the toolbar above the database explorer.
Right click on the Subject01 > Review raw file:
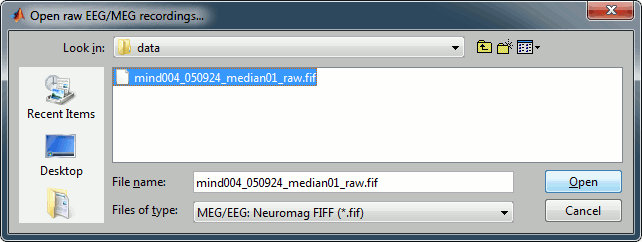
Select the file format: MEG/EEG: Elekta-Neuromag (*.fif)
Pick the file: sample_neuromag/data/mind004_050924_median01_raw.fif
Select "Event channel" when asked about the events.
Then you have to select from which technical tracks you want to read the events.
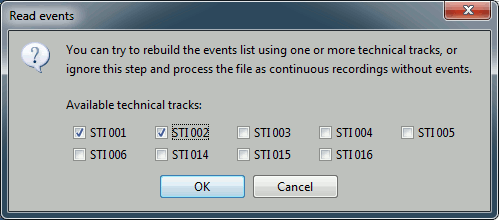
- In this dataset, the tracks of interest are:
STI 001: Contains the onsets of the electric stimulation of the right arm.
STI 002: Contains the onsets of the electric stimulation of the left arm.
- These are not standard settings, they depend on your acquisition setup. In most cases, you would rather import "events" defined with the Neuromag acquisition software, which are saved in tracks STI 014, STI 101, STI 201...
Select STI 001 and STI 002. The full tracks are read, all the changes of values are detected from these two tracks and a list of events is created.
At the end, you're asked if you want to "Refine the registration with the head points". This operation improves the initial MRI/MEG registration by fitting the head points digitized at the MEG acquisition and the scalp surface. Answer YES.
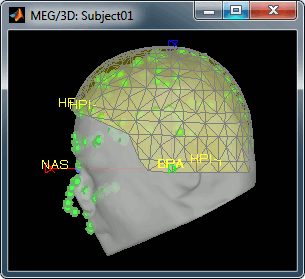
Always make sure that the final registration looks good. The yellow surface represents the inner surface of the MEG helmet and the green dots represent the head points that were saved with the MEG recordings. In this case the registration looks acceptable. If you consider it is not the case, you can fix it manually: right-click on the channel file > MRI registration > Edit.
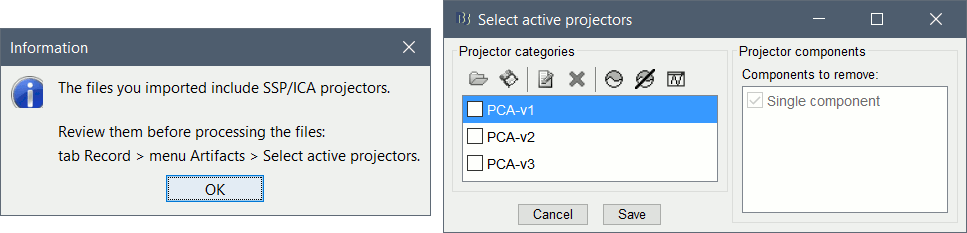
A message will finally indicate that some default SSP projectors are available in the file. This is documented in an advanced section at the end of this page. We will not use them here, simply close the window "Select active projectors".
Review the recordings
Right-click on the "Link to raw file" to see what types of channels are available:
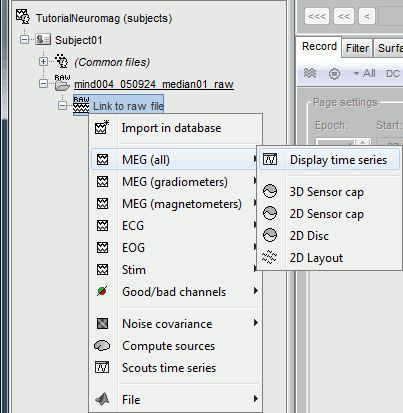
MEG (all): Represents all the MEG sensors in the file (magnetometers and gradiometers)
MEG (magnetometers): 101 magnetometers (sensor names ending with a "1")
MEG (gradiometers): 202 planar gradiometers (sensor names ending with "2" or "3")
ECG: Electro-cardiogram
EOG: Electro-oculogram
Stim: All the stimulation lines
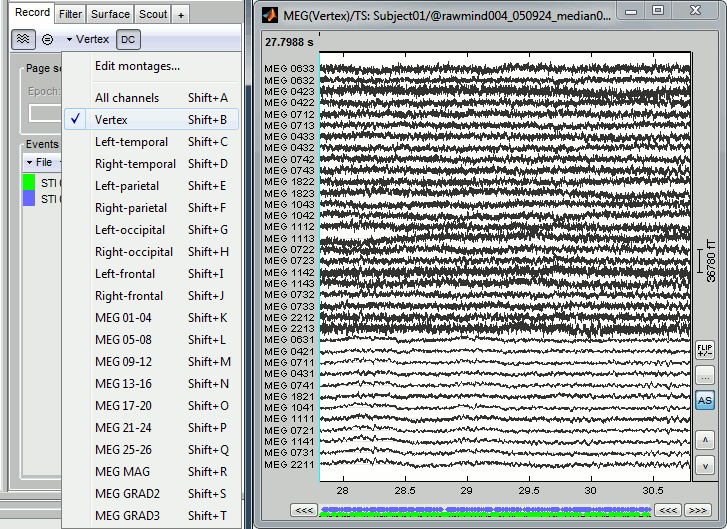
- Double-click on the file to display all the MEG sensors.
In the Record tab, look at the list of the available default display configurations for the Elekta-Neuromag Vectorview MEG systems. If you would like to use different combinations of sensors, you can create your own combinations with the entry Edit montage.
Pre-processing
Evaluate the recordings
- Drag and drop the "Link to raw file" into the Process1 list.
Select the process "Frequency > Power spectrum density", configure it as follows:
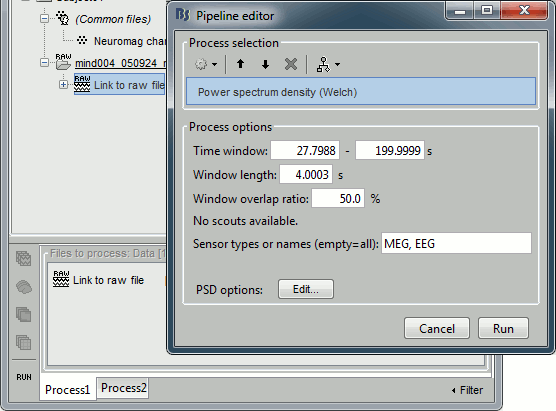
- In all the processes, the types of sensors can be specified using the following convention:
MEG: All the MEG sensors
MEG MAG: All the magnetometers
MEG GRAD: All the gradiometers
MEG GRAD2: Only the first axial gradiometer on each chip (name ends with a "2")
MEG GRAD3: Only the second axial gradiometer on each chip (name ends with a "3")
Double-click on the PSD file to display it.
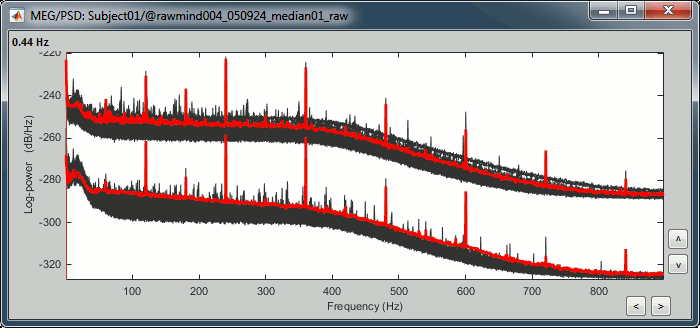
The sensors are split in two groups because the two types of MEG sensors record different ranges of values: the gradiometer values are always higher than the magnetometers.
- This power spectrum shows that there is no MEG sensor that is clearly bad, but some sensors show a strong contamination from the power lines (60Hz and harmonics).
Note that if you use the continuous head tracking capabilities of your Elekta system, you would see a lot more noise due to this HPI coil activity in the PSD. This is illustrated in this other tutorial.
Remove: 60Hz and harmonics
Warning: This process is very time and resource consuming. If you are not planning on analyzing anything specifically around 60Hz, you may want to skip this section and the long computation time associated with it. It is described here as a reference for users who would need it.
Select the process "Pre-process > Notch filter", configure it as follows:
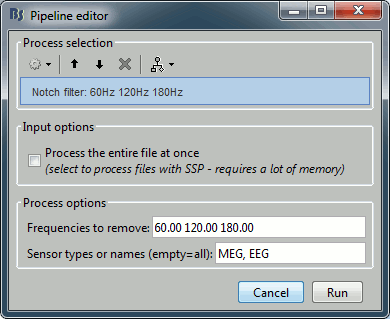
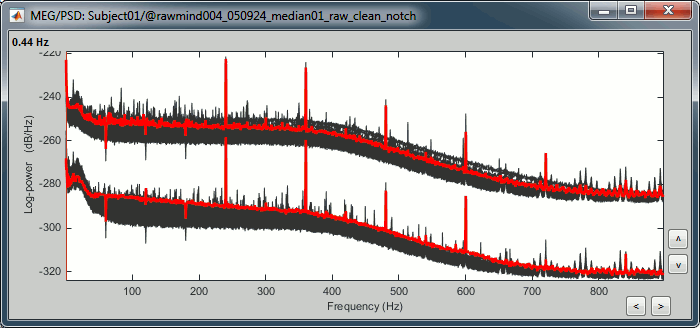
To evaluate the results of this process, select the new file ("Raw | notch") and run again the process "Frequency > Power spectrum density".
You should observe a significant decrease of the contributions of the removed frequencies (60Hz, 120Hz, 180Hz) compared with the original spectrum.
Heartbeats and blinks
Signal Space Projection (SSP) is a method for projecting the recordings away from stereotyped artifacts, such as eye blinks and heartbeats.
Double-click on the filtered continuous file to display all the MEG recordings.
Right-click on the link > ECG > Display time series, to look at the heartbeats.
Right-click on the link > EOG > Display time series, to look at the eye movements.
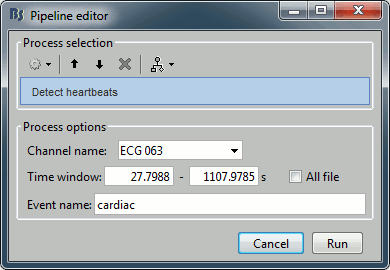
- From the Artifacts menu in the Record tab, run the detection processes:
Artifacts > Detect heartbeats: Select channel ECG 063, event name "cardiac".
Artifacts > Detect eye blinks: Select channel EOG 062, event name "blink".
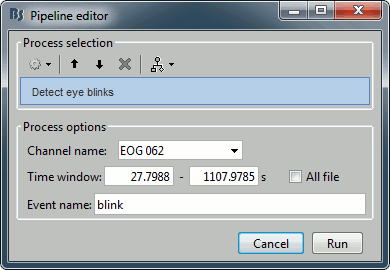
Artifacts > Remove simultaneous: To avoid capturing ocular artifacts in the cardiac SSP.
Remove events named: "cardiac"
When too close from events: "blink"
Minimum delay between events: 250ms.
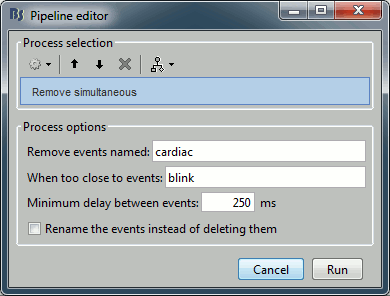
Review the traces of ECG/EOG channels and make sure the events detected make sense.
Artifacts > SSP: Heartbeats: Event "cardiac", sensors="MEG MAG", use existing SSP.
(select component #1)
Artifacts > SSP: Heartbeats: Event "cardiac", sensors="MEG GRAD", use existing SSP.
(select component #1)
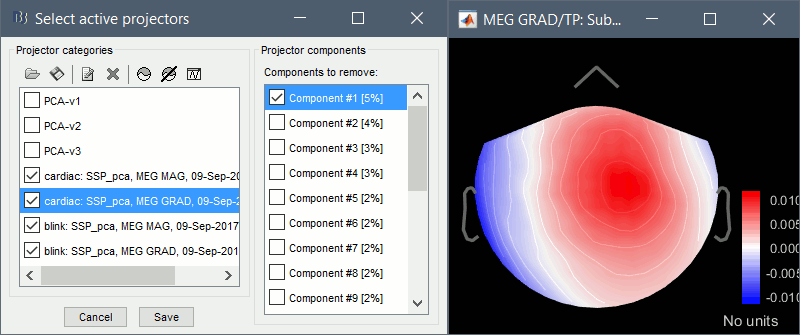
Artifacts > SSP: Eyeblinks: Event "blink", sensors="MEG MAG", use existing SSP.
(select component #1)
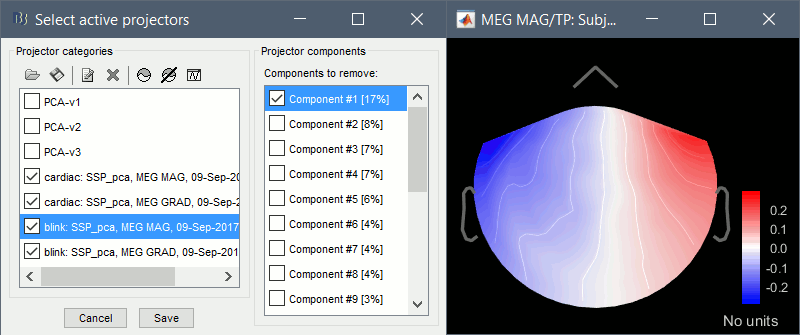
Artifacts > SSP: Eyeblinks: Event "blink", sensors="MEG GRAD", use existing SSP.
(select component #1)
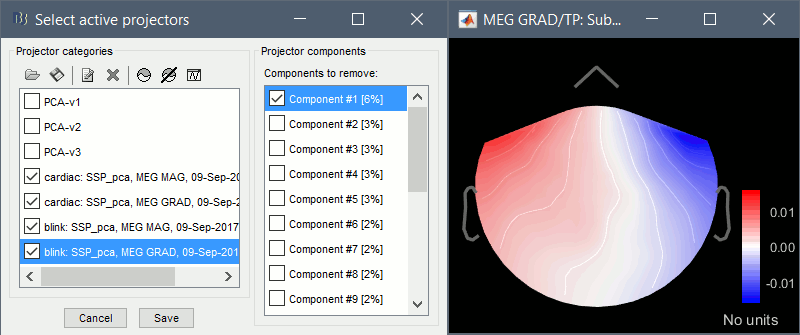
For more information regarding this method, refer to the tutorial: Artifact cleaning with SSP.
At this stage, you should also review the entire file (pressing repeatedly F3) to detect any obvious bad segment in the recordings. If you detect any, select the time window to mark as bad, right click on the figure > Reject time segment.
You could also use the automatic detection for other artifacts: Additional bad segments.
Epoching and averaging
Import the recordings
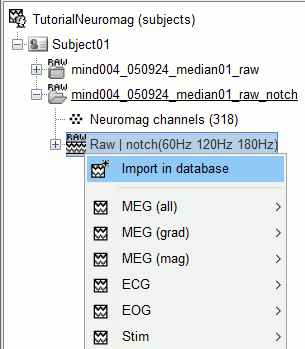
Right-click on the raw link > Import in database.
The following figure appears, that asks how to import these recordings in the Brainstorm database.
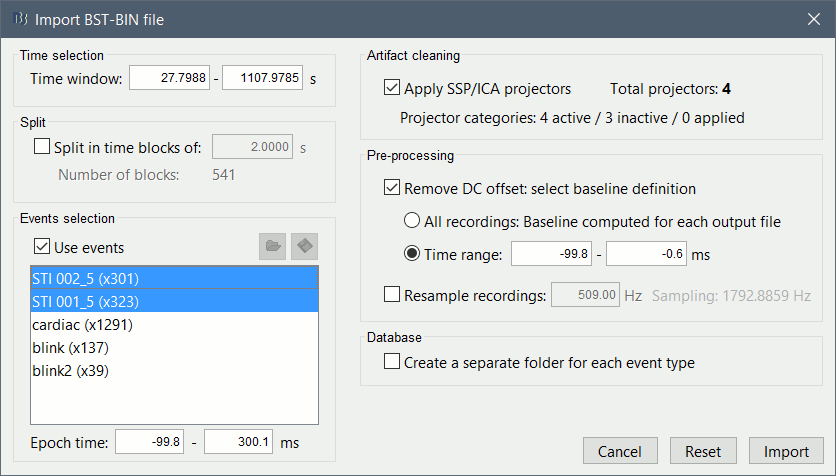
Time window: Time range of interest, keep all the time definition.
Split: Useful to import continuous recordings without events. We do not need this here.
Events selection: Check the "Use events" option, and select both STI 001 and STI 002. The values in the parenthesis represent the number of occurrences of each event.
Epoch time: Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms
Use Signal Space Projections: Save and use the SSP vectors created by MaxFilter, MNE and Brainstorm. Keep this option selected.
Remove DC Offset: Check this option, and select: Time range: [-100, 0] ms.
For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), then subtract it from the channel at all the times in [-100,+300]ms.Resample recordings: Keep this unchecked
Create a separate folder for each epoch type: If selected, a new folder is created for each event type (here, it would create two folders "STI 001" and "STI 002"). This option is mostly for EEG studies with channel files shared across runs. In a MEG study, we usually recommend to use one channel file per run, and to import all the epochs from one run in the same folder.
- Click on [Import] and wait.
Rename the two groups of trials "STI_001_5" and "STI_002_5" into respectively "Right" and "Left" (Right-click > Rename, or press F2, or click twice waiting a while before the two clicks).

At this stage, you should review all the trials (press F3 to jump to the next file), separately for the magnetometers and the gradiometers, to make sure that you don't have any bad trial that have been imported. If you find a bad trial: right-click on the file or on the figure > Reject trial.
Averaging
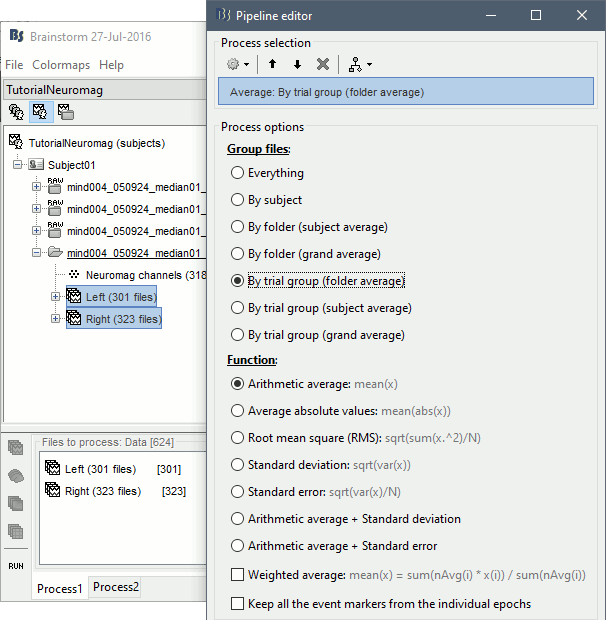
- Drag and drop all the left and right trials to the Process1 tab.
Run the process Average > Average files > By trial group (folder average):
Double-click on the Left and Right averages to display all the MEG sensors:
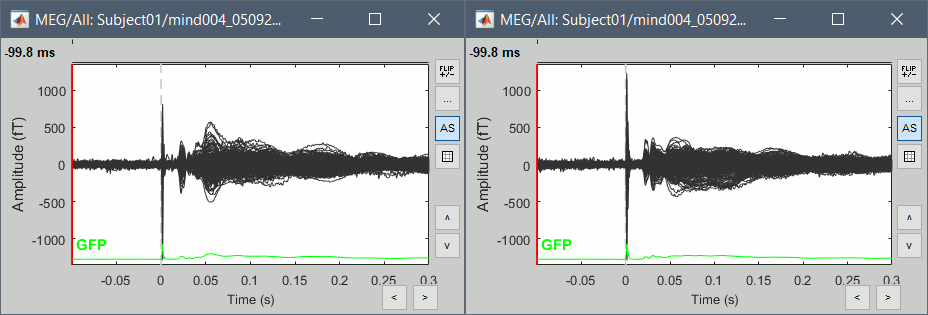
Stimulation artifact
- We see clearly our evoked responses in these figures, but the stimulation artifact between 0ms and 5ms causes the automatic scaling of the figures to be too high, especially for the right stimulation. This effect would get a lot worse when displaying the sources for these files. To get a better display of these averages, we can cut these peaks at 0ms by replacing the values with linear interpolations.
- Select the averages for the Left and Right condition in the Process1 tab.
Run the process Artifacts > Cut stimulation artifact: 0-5ms, overwrite
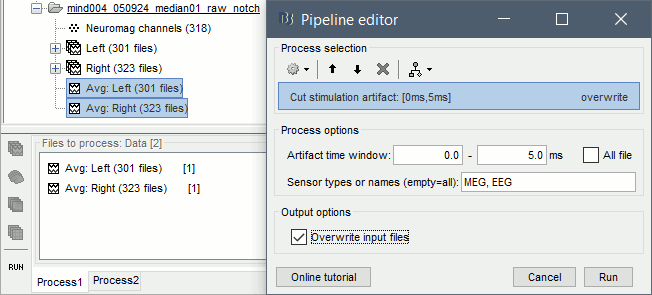
Now you can display the averages correctly: Right-click > MEG (All) > Display time series.
Display the 2D topographies for each average: Right-click on the file > MEG (All) > 2D Sensor cap (or press CTRL+T from the time series figures).
You can also add a low-pass filter at 100Hz for the visualization.
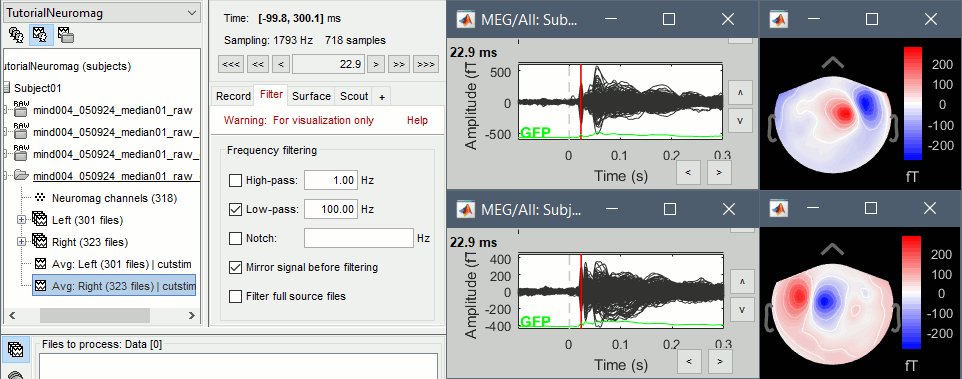
- Everything looks ok, the time series figures show a clean signal with a very high SNR, and the topographies at the first response peak (about 22ms) show an activity on the left side for the Right condition, and on the right side for the Left condition.
Gradiometers & magnetometers
Scaling factor
Magnetometers record values in Tesla (T), while gradiometers record values in Tesla per meter (T/m). The range of values obtained is not the same, therefore it is difficult to represent the two types of signals in the same figures.
We can convert the gradiometers values from T/m to T by multiplying them with the distance between the two gradiometer coils (0.0168 meters on the VectorView MEG system). But this still produces values that are too small to be represented with the same scale as the magnetometers. A more practical multiplication factor of 0.04 was proposed by Matti Hamalainen.
In the time series figure and for Elekta-Neuromag systems (all versions), the gradiometer values are always multiplied by 0.04. This is an empirical scaling factor that is used for visualization only, it is never saved in the recordings and is not used for any other purpose. See function bst_scale_gradmag.m.
Magnetic interpolation
You can also display the MEG MAG (top) and MEG GRAD (bottom) sensors separately:
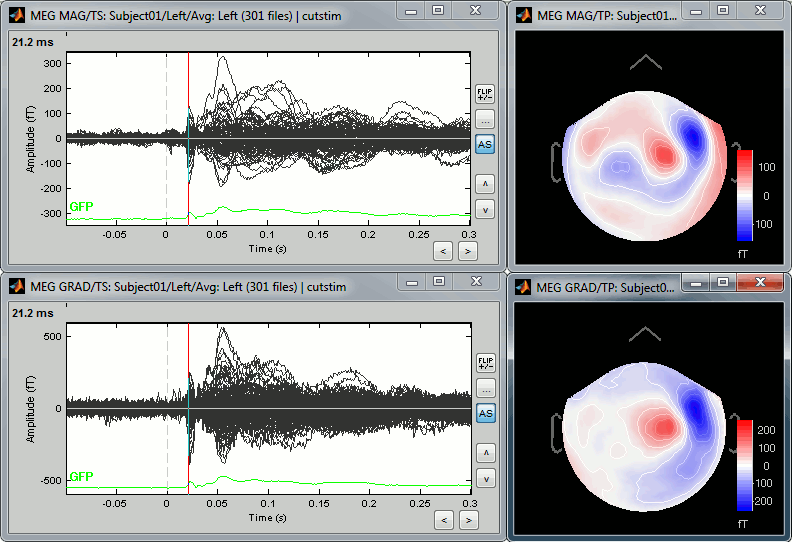
- The topographies look very similar, because all the values are re-interpolated on a surface of virtual magnetometers. To display the real values for the different types of sensors, without this magnetic interpolation, use the menus "No magnetic interpolation".
Right-click on Left > MEG (mag) > No magnetic interpolation > 2D Sensor cap.
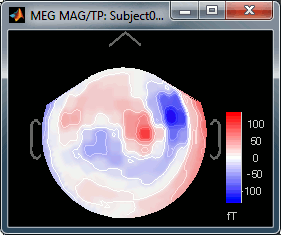
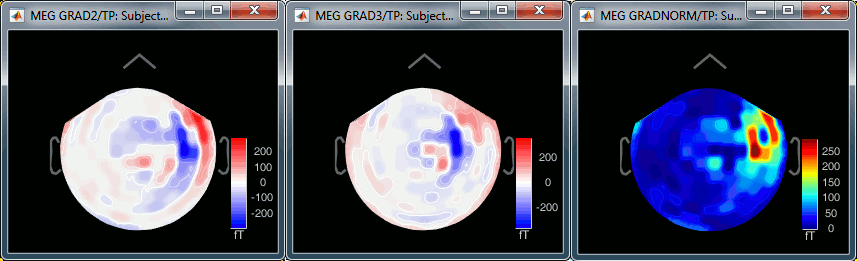
- Repeat the same operation for "MEG (gradiometers)". You now have three options, because it is not possible to display at the same time the values for the two planar gradiometers:
Grad 2: First gradiometer, sensor names ending with "2"
Grad 3: Second gradiometer, sensor names ending with "3"
Grad norm: Norm of the two gradiometers => sqrt(grad22 + grad32)
The three options side-by-side at 22ms (grad 2, grad3, norm):
Source estimation
We need now to calculate a source model for these recordings, using a noise covariance matrix calculated from the pre-stimulation baselines. This process is not detailed a lot here because it is very similar to what is shown in the introduction tutorials.
Head model
Right-click on the channel file > Compute head model. Keep the default options.
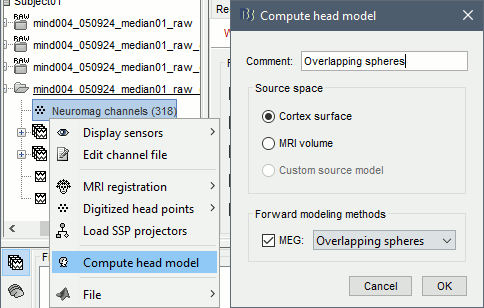
For more information: Head model tutorial.
Noise covariance
Select all the Left and Right trials, right-click > Noise covariance > Compute from recordings.
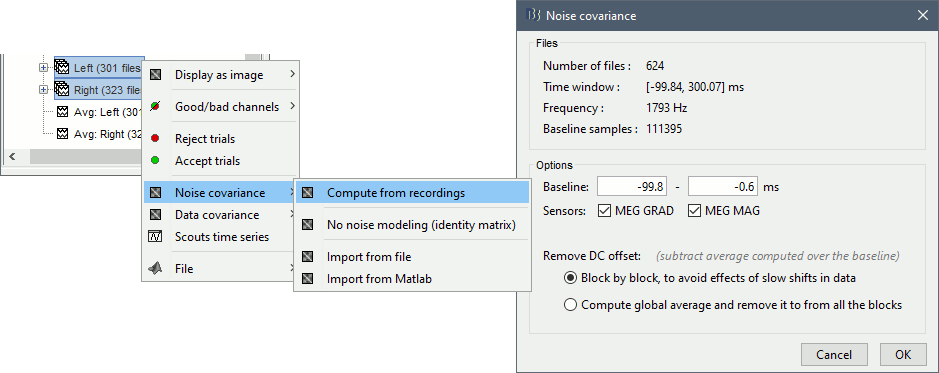
- Leave all the default options and click [OK].
For more information: Noise covariance tutorial.
Inverse model
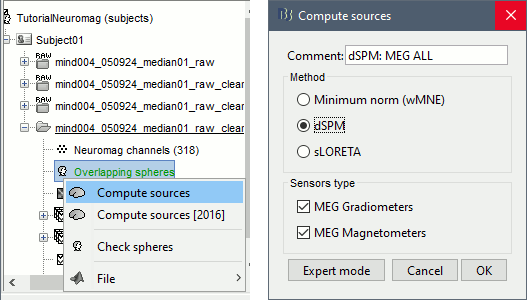
Right-click on the headmodel > Compute sources. Select dSPM and all the default options. This will run a source reconstruction grouping the information from the magnetometers and the gradiometers into the same model. This is the recommended procedure, but you can also estimate sources for the two types of sensors separately if needed.
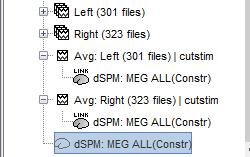
- A shared inversion kernel is created in the same folder, and a link node is now visible for each recording file, single trials and averages.
For more information: Source estimation tutorial.
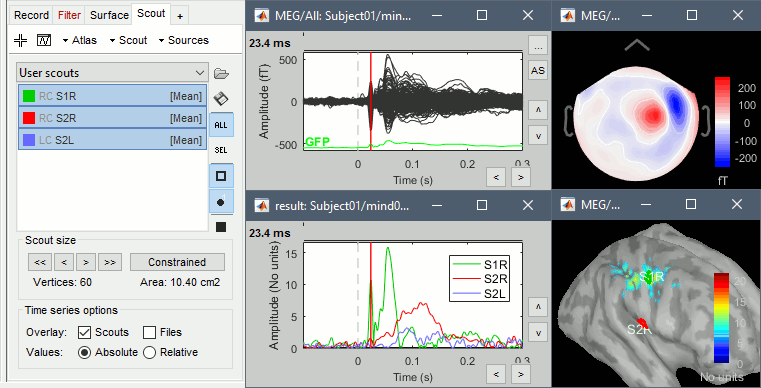
Scouts
Place scouts to capture the activity in the primary and secondary somatosensory areas to track the processing of the electric stimulations in time, at the surface surface of the brain.
Existing SSP projectors
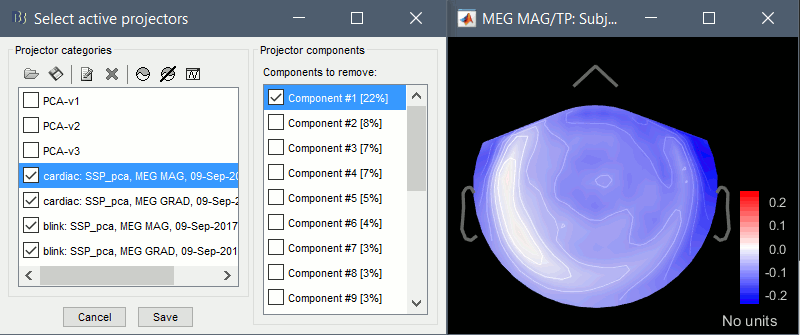
In the Record tab, select menu "Artifacts > Select active projectors". You will see the file contains three SSP projectors that are not applied by default.
In general, every Neuromag Vectorview (306 channel) system has at least two default SSP files, one for the supine position and one for upright. The newer Triux machines may contain a third SSP set. The default bases for the Vectorview are rank 5 for planar gradiometers, and rank 8 for magnetometers, for 13 bases total, but applied separately to the two types of sensors. Some sites may have different rank default bases, such as the massive three-layer magnetically shielded rooms, which may use even smaller rank bases sets.
During acquisition, the real-time display projects the data away from these SSP vectors, for display purposes only. If U is the SSP orthonormal basis, then the data are displayed as (d - U U^T d). The original data "d" are stored without the SSP applied, along with all of the default SSP vectors, U.
The default SSP bases are calculated from an empty room recording, often as part of the initial installation of the site, and they rarely need changing, since they are essentially formed by the shape of the room in its environment, and by the position of the instrument within this room, hence the need for different bases dependent on the position of the helmet.
If a subject has a very noisy low frequency baseline disturbance, such as due to metal artifacts on the patient, it is beneficial to recalculate SSP bases from a short period of their data (e.g. 2 minutes). Matti Hamalainen's MNE program can regenerate valid SSP data files compatible with the Vectorview, and these can be used in place of the default SSP bases, after restarting the acquisition program. These custom SSP bases are effective for allowing the technician acquiring the data to confirm that all channels continue to acquire usuable data. The machine can then be reset to the defaults. All of these actions can be effected through relatively simple scripts that regenerate the SSP bases, using MNE, in near real-time.
After the data are acquired, the original SSP bases are often ignored in subsequent processing. They can be useful for initial viewing of the data. For research analyses, rather than use the default SSP bases, it is preferential to recalculate the noise covariance from appropriate baselines, to include brain noise if possible, and use that as the full noise covariance. SSP is essentially an inverted noise covariance with the eigenvalues set to either 0 (reject) or 1 (pass), which is a generally inadequate description of the noise.
At many sites, the MaxFilter "TSSS" program is typically run, which is an implicit LCMV beamformer (programmed as a Generalized Sidelobe Canceller), which has the benefit that it implicitly whitens a lot of the background noise as part of its processing. Therefore, MaxFilter also does not use the original SSP bases, and as part of its default operation, MaxFilter always deletes the original SSP vectors and does not include them in the output FIF file. (As an aside, if motion compensation is on, then MaxFilter adds nine synthetic channels to the data to track head motion, written out using quaternions). Thus when you load data processed by MaxFilter, you will not find any of the original SSP projectors.
This topic is also discussed on the forum: Neuromag autossp
For additional information about SSP, read the tutorial: Artifact cleaning with SSP.
Existing SSP and pre-processing
If you select the default SSP projectors and try to run the process "Notch filter" without selecting the option "Process the entire file at once", you would get the following error. Using these projectors requires all the sensors, and filtering requires all the time samples at once. As a consequence, the entire file must be read in one block in order to apply frequency filters on it.
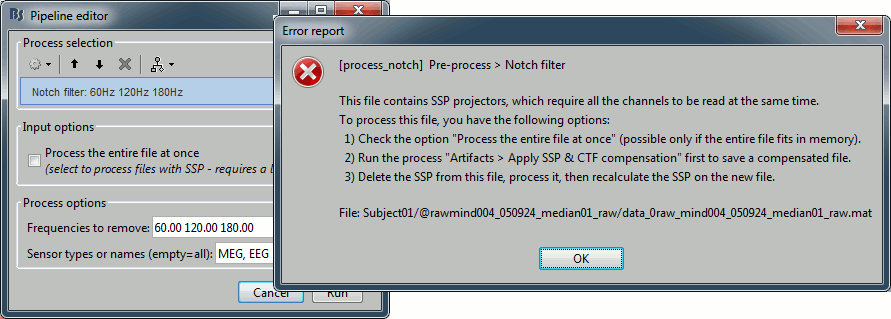
Alternatively, if you try with the option "Process the entire file at once", you would probably crash Matlab or freeze your computer because of a lack of memory. The file is already large and this process requires to create a lot of matrices of the same size in memory. Do not try this unless you have at least 24Gb of memory available on your computer.
The solution is to re-write an intermediate file with the SSP projectors applied, that you will be able to process easily after. In Process1, keep the "Link to raw file" selected.
Run process "Artifacts > Apply SSP and CTF compensation". This creates a new continuous file which contains the same data as the original .fif file, but with the SSP projectors applied to the recordings values (instead of being just saved as linear operators on the side).
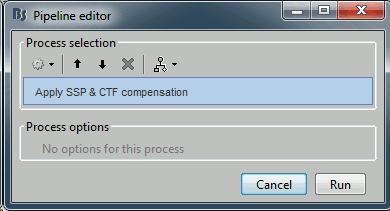
Clear the Process1 list and select the new continuous file "Raw | clean"
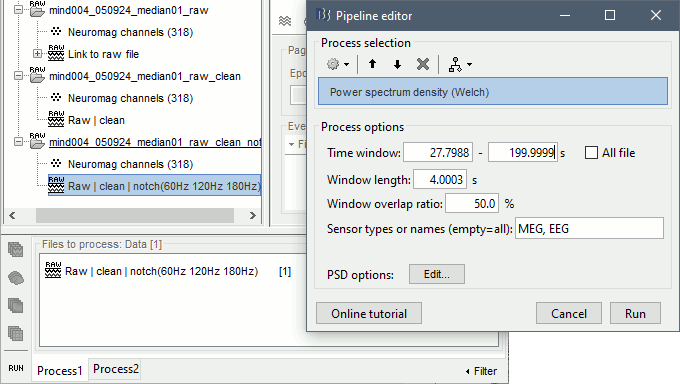
Run the process "Pre-process > Notch filter", configured as follows:

To evaluate the results of this process, select the last file generated ("Raw | clean | notch") and run again the process "Frequency > Power spectrum density".
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_neuromag.m
Discussion
Combining magneto+gradiometers: http://neuroimage.usc.edu/forums/showthread.php?1900
Additional Reading
Weisend MP, Hanlon FM, Montano R, Ahlfors SP, Leuthold AC, Pantazis D, Mosher JC, Georgopoulos AP, Hämäläinen MS, Aine CJ. "Paving the way for cross-site pooling of magnetoencephalography (MEG) data." InInternational Congress Series 2007 Jun 1 (Vol. 1300, pp. 615-618). Elsevier. https://doi.org/10.1016/j.ics.2006.12.095