MEG visual tutorial: Group analysis (BIDS)
Authors: Francois Tadel, Elizabeth Bock.
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "Multimodal, Multisubject data fusion".
It is part of a collective effort to document and standardize MEG/EEG group analysis, see Frontier's research topic: From raw MEG/EEG to publication: how to perform MEG/EEG group analysis with free academic software.
The data processed here consists in simultaneous MEG/EEG recordings of 16 subjects performing a simple visual task on a large number of famous, unfamiliar and scrambled faces. This tutorial follows another page that explains how to process one single subject in details.
License
This dataset was obtained from the OpenNeuro project (https://openneuro.org/), accession #ds117. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset, Scientific Data (2015)
Any questions, please contact: rik.henson@mrc-cbu.cam.ac.uk
For citing the analysis, the processing pipeline is published in this article:
Tadel F, Bock E, Niso G, Mosher JC, Cousineau M, Pantazis D, Leahy RM, Baillet S, MEG/EEG Group Analysis With Brainstorm, Frontiers in Neuroscience, Feb 2019
Download and installation
First, make sure you have enough space on your hard drive, at least 40Gb:
The Brainstorm database with all the data imported, downloaded from this website: 20Gb
The same database after this tutorial: 40Gb
You can follow this tutorial after processing the recordings for the 16 good subjects (6 runs per subject) as illustrated in the single subject tutorial. Otherwise, we provide a Brainstorm protocol that includes all the imported data, downsampled at 275Hz:
Go to the Download page, download the file TutorialGroup.zip (20Gb).
- Unzip this file in your Brainstorm database folder (brainstorm_db).
In Brainstorm, menu File > Load protocol > Load from folder > Select brainstorm_db/TutorialGroup
The database you need in order to follow this tutorial should contain the following:
The individual anatomy imported from FreeSurfer for each subject (16 subjects).
- The sensor level averages (MEG+EEG) for each run (downsampled to 275Hz or not).
The forward model, noise covariance and inverse models for each subject and each run.
This protocol TutorialGroup is produced from the single subject protocol TutorialVisual with the script: brainstorm3/toolbox/script/tutorial_visual_copy.m
1 function tutorial_visual_copy(ProtocolNameSingle, ProtocolNameGroup, reports_dir)
2 % TUTORIAL_VISUAL_COPY: Copy the subject averages for the Brainstorm/SPM group tutorial into a new protocol (BIDS VERSION)
3 %
4 % ONLINE TUTORIALS:
5 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle
6 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup
7 %
8 % INPUTS:
9 % - ProtocolNameSingle : Name of the protocol created with all the data imported (TutorialVisual)
10 % - ProtocolNameGroup : Name of the protocol with just the averages, downsampled to 275Hz (TutorialGroup)
11 % - reports_dir : If defined, exports all the reports as HTML to this folder
12
13 % @=============================================================================
14 % This function is part of the Brainstorm software:
15 % https://neuroimage.usc.edu/brainstorm
16 %
17 % Copyright (c) University of Southern California & McGill University
18 % This software is distributed under the terms of the GNU General Public License
19 % as published by the Free Software Foundation. Further details on the GPLv3
20 % license can be found at http://www.gnu.org/copyleft/gpl.html.
21 %
22 % FOR RESEARCH PURPOSES ONLY. THE SOFTWARE IS PROVIDED "AS IS," AND THE
23 % UNIVERSITY OF SOUTHERN CALIFORNIA AND ITS COLLABORATORS DO NOT MAKE ANY
24 % WARRANTY, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO WARRANTIES OF
25 % MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE, NOR DO THEY ASSUME ANY
26 % LIABILITY OR RESPONSIBILITY FOR THE USE OF THIS SOFTWARE.
27 %
28 % For more information type "brainstorm license" at command prompt.
29 % =============================================================================@
30 %
31 % Author: Francois Tadel, Elizabeth Bock, 2016-2018
32
33 % ===== CHECK PROTOCOLS =====
34 % Start brainstorm without the GUI
35 if ~brainstorm('status')
36 brainstorm nogui
37 end
38 % Output folder for reports
39 if (nargin < 3) || isempty(reports_dir) || ~isdir(reports_dir)
40 reports_dir = [];
41 end
42 % You have to specify the folder in which the tutorial dataset is unzipped
43 if (nargin < 2) || isempty(ProtocolNameSingle) || isempty(ProtocolNameGroup)
44 ProtocolNameSingle = 'TutorialVisual';
45 ProtocolNameGroup = 'TutorialGroup';
46 end
47
48 % Output protocol: Delete existing protocol
49 gui_brainstorm('DeleteProtocol', ProtocolNameGroup);
50 % Output protocol: Create new protocol
51 iProtocolGroup = gui_brainstorm('CreateProtocol', ProtocolNameGroup, 0, 0);
52 % Output protocol: Get protocol information
53 ProtocolInfoGroup = bst_get('ProtocolInfo');
54
55 % Input protocol: Check that it exists
56 iProtocolSingle = bst_get('Protocol', ProtocolNameSingle);
57 if isempty(iProtocolSingle)
58 error(['Unknown protocol: ' ProtocolNameSingle]);
59 end
60 % Select input protocol
61 gui_brainstorm('SetCurrentProtocol', iProtocolSingle);
62 % Input protocol: Get protocol information
63 ProtocolInfoSingle = bst_get('ProtocolInfo');
64
65
66 % ===== COPY ONLY GOOD SUBJECTS =====
67 % Start a new report (one report per subject)
68 bst_report('Start');
69 % Loop on subjects
70 for iSubj = 1:16
71 % Subject folders
72 SubjectName = sprintf('sub-%02d', iSubj);
73 AnatSrc = bst_fullfile(ProtocolInfoSingle.SUBJECTS, SubjectName);
74 DataSrc = bst_fullfile(ProtocolInfoSingle.STUDIES, SubjectName);
75 AnatDest = bst_fullfile(ProtocolInfoGroup.SUBJECTS, SubjectName);
76 DataDest = bst_fullfile(ProtocolInfoGroup.STUDIES, SubjectName);
77 % If subject folder doesn't exist: skip
78 if ~file_exist(AnatSrc) || ~file_exist(DataSrc)
79 disp(['Subject "' SubjectName '" does not exist or is incomplete.']);
80 continue;
81 end
82 % Copy anatomy files
83 mkdir(AnatDest);
84 disp(['Copying: ' AnatSrc ' to ' AnatDest '...']);
85 file_copy(bst_fullfile(AnatSrc, '*.mat'), AnatDest);
86 % Copy analysis folders
87 mkdir(bst_fullfile(DataDest, '@default_study'));
88 mkdir(bst_fullfile(DataDest, '@intra'));
89 disp(['Copying: ' DataSrc ' to ' DataDest '...']);
90 file_copy(bst_fullfile(DataSrc, '@default_study', '*.mat'), bst_fullfile(DataDest, '@default_study'));
91 file_copy(bst_fullfile(DataSrc, '@intra', '*.mat'), bst_fullfile(DataDest, '@intra'));
92 % Loop on runs
93 for iRun = 1:6
94 % Run folders
95 RunName = sprintf('sub-%02d_ses-meg_task-facerecognition_run-%02d_proc-sss_meg_notch', iSubj, iRun);
96 RunSrc = bst_fullfile(DataSrc, RunName);
97 RunDest = bst_fullfile(DataDest, RunName);
98 % If run folder doesn't exist: skip
99 if ~file_exist(RunSrc)
100 disp(['Run "' SubjectName '/' RunName '" does not exist or is incomplete.']);
101 continue;
102 end
103 % Copy files
104 mkdir(RunDest);
105 disp(['Copying: ' RunSrc ' to ' RunDest '...']);
106 file_copy(bst_fullfile(RunSrc, 'brainstormstudy.mat'), RunDest);
107 file_copy(bst_fullfile(RunSrc, 'channel_*.mat'), RunDest);
108 file_copy(bst_fullfile(RunSrc, '*_average_*.mat'), RunDest);
109 if ~isempty(dir(bst_fullfile(RunSrc, 'headmodel_*.mat')))
110 file_copy(bst_fullfile(RunSrc, 'headmodel_*.mat'), RunDest);
111 end
112 if ~isempty(dir(bst_fullfile(RunSrc, 'noisecov_full.mat')))
113 file_copy(bst_fullfile(RunSrc, 'noisecov_full.mat'), RunDest);
114 end
115 if ~isempty(dir(bst_fullfile(RunSrc, 'results_*.mat')))
116 file_copy(bst_fullfile(RunSrc, 'results_*.mat'), RunDest);
117 end
118 if ~isempty(dir(bst_fullfile(RunSrc, 'timefreq_*.mat')))
119 file_copy(bst_fullfile(RunSrc, 'timefreq_*.mat'), RunDest);
120 end
121 end
122 end
123
124 % ===== RELOAD =====
125 % Reload output protocol
126 db_reload_database(iProtocolGroup);
127 % Select output protocol
128 gui_brainstorm('SetCurrentProtocol', iProtocolGroup);
129
130 % ===== DOWNSAMPLE TO 275HZ =====
131 % Process: Select data files in: */*
132 sDataAll = bst_process('CallProcess', 'process_select_files_data', [], []);
133 % Process: Resample: 275Hz
134 sDataAll = bst_process('CallProcess', 'process_resample', sDataAll, [], ...
135 'freq', 275, ...
136 'overwrite', 1);
137 % Process: Select time-frequency files in: */*
138 sTimefreqAll = bst_process('CallProcess', 'process_select_files_timefreq', [], []);
139 % Process: Resample: 275Hz
140 if ~isempty(sTimefreqAll)
141 sTimefreqAll = bst_process('CallProcess', 'process_resample', sTimefreqAll, [], ...
142 'freq', 275, ...
143 'overwrite', 1);
144 end
145
146 % ===== RENAME: DATA =====
147 % Process: Select data files in: */*
148 sDataAll = bst_process('CallProcess', 'process_select_files_data', [], []);
149 % Rename data files
150 for i = 1:length(sDataAll)
151 % Remove all the processing tags
152 iTag = strfind(sDataAll(i).Comment, ' |');
153 if isempty(iTag)
154 continue;
155 end
156 newComment = sDataAll(i).Comment(1:iTag-1);
157 % Process: Set comment: AA
158 bst_process('CallProcess', 'process_set_comment', sDataAll(i), [], ...
159 'tag', newComment, ...
160 'isindex', 0);
161 end
162
163 % ===== RENAME: TIME-FREQ =====
164 % Process: Select time-frequency files in: */*
165 sTimefreqAll = bst_process('CallProcess', 'process_select_files_timefreq', [], []);
166 % Rename timefreq files
167 %AllConditions = {'Famous', 'Scrambled', 'Unfamiliar'};
168 for i = 1:length(sTimefreqAll)
169 % Remove all the processing tags
170 iTag = strfind(sTimefreqAll(i).Comment, ' |');
171 if isempty(iTag)
172 continue;
173 end
174 newComment = sTimefreqAll(i).Comment(1:iTag-1);
175 %newComment = ['Avg: ', AllConditions{sTimefreqAll(i).iItem}, ', Power, 6-60Hz'];
176 % Process: Set comment
177 bst_process('CallProcess', 'process_set_comment', sTimefreqAll(i), [], ...
178 'tag', newComment, ...
179 'isindex', 0);
180 end
181
182 % Save report
183 ReportFile = bst_report('Save', []);
184 if ~isempty(reports_dir) && ~isempty(ReportFile)
185 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolNameGroup '_copy.html']));
186 end
187
188
189
190
191
Overview of the analysis
Coregistration of the acquisition runs
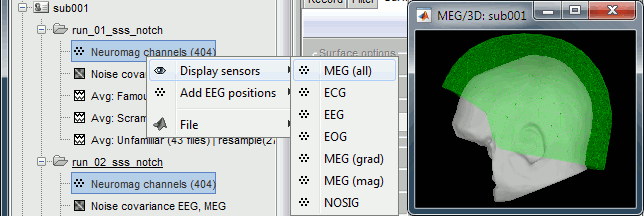
For each subject, all the runs have been registered to a common head position with MaxFilter. To verify this, select all the channel files within one subject, right-click > Display sensors > MEG (all). The surfaces representing the MEG helmet are perfectly overlapping for all the runs. When the runs are not aligned, it looks like this.
This means we can safely average or compare the MEG sensor values across runs within one subject. However, it is not reliable to average MEG recordings across subjects, because of the anatomical differences between subjects.
This doesn't mean we can estimate the sources only once per subject. We have computed different SSP projectors and selected different bad channels for each acquisition run. To be able to use this information efficiently we should estimate the sources for each run separately, then average the sources across runs.
The forward model is the same for all the runs within one subject, therefore it can be computed for the first run and copied to all the other runs.
Objectives
The objectives for this tutorial are to reproduce the analysis presented in the following documents:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset
Scientific Data (2015)
Wakeman DG, Henson RN, Functional and structural connrctivity in face processing: MEG, EEG, fMRI, MRI and DWI data, Biomag 2016
SPM12 manual, chapter 41: Multimodal, Multisubject data fusion
Summary of the results we will compute in this tutorial:
- Subject and grand averages for each condition (Famous, Unfamiliar, Scrambled).
- Normalization of these averages (Z-score for the sources, or ERS/D for the time-frequency maps).
- Projection of the sources results on a template and spatial smoothing of the source maps.
- Contrast between faces (famous+unfamiliar) and non-faces (scrambled): difference of averages and significance test.
- Contrast between famous faces and unfamiliar faces: difference of averages and significance test.
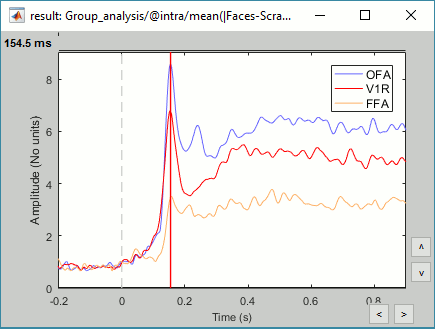
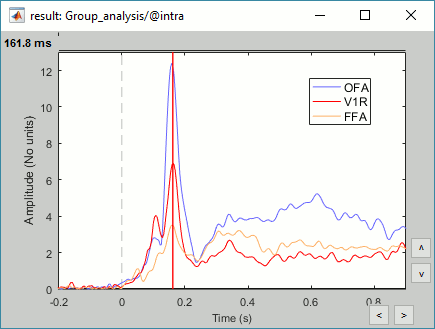
- Sensors of interest: EEG070 (or EEG060 or EEG065 if EEG070 is marked as bad)
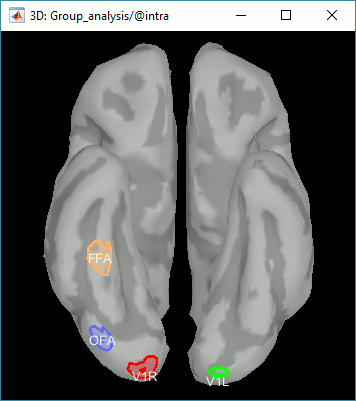
Expected effects:
- OFA: Bilateral occipital face area
- FFA: Bilateral fusiform face area
- STS: Right superior temporal sulcus
- Around 170ms (N170): Difference between Faces and Scrambled.
- After 250ms: Difference between Famous and Unfamiliar.
The methodology that we will follow for computing the averages and the other statistics is described in the tutorial "Workflows".
This tutorial page illustrates how to run the analysis using the interface. In practice, with a larger number of subjects, going through all these steps manually with the Brainstorm GUI is not practical, time consuming and prone to manipulation errors. We encourage our users to script their group analyses instead of performing all these steps manually. Typically, one would select a few subjects, prototype the group analysis pipeline with the interactive interface, generate the corresponding Matlab script, and finally run it on all the subjects of the database in reproducible way. Assembling a processing script is discussed in the online tutorial Scripting and illustrated with the scripts tutorial_frontiers2018*.m.
Subject averages: Famous, Unfamiliar, Scrambled
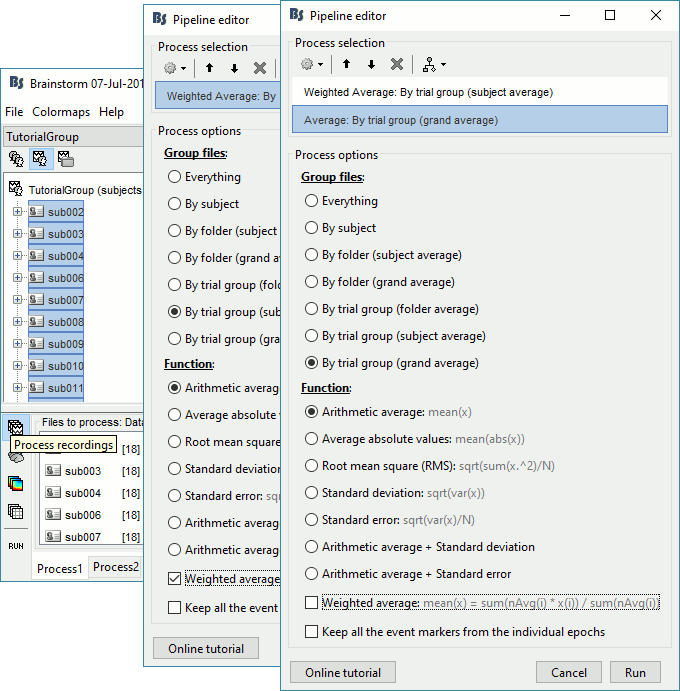
We will start by computing the subject-level averages for all the data types we have: sensor-level recordings, source maps and time-frequency results. We will use a weighted average to group the results we have for each run (weighted by the number of good trials). We want to compute the averages for each experimental condition separately (famous, unfamiliar, scrambled).
Since the files are already selected in the interface, we will also compute a grand average across subject. In this context, we will consider that each subject has the same weight in the grand average (option "weighted" not selected). Note that it is not accurate to average MEG recordings across subjects, but we can do it just to get a general idea of the group effects (more information).
MEG/EEG
Drag and drop all the subjects in Process1. Select button [Process recordings].
Select process Average > Average files: By trial group (subject average)
Arithmetic average, Weighted
Add process Average > Average files: By trial group (grand average)
Arithmetic average, Not weighted
In output, you get three grand averages in the "Group analysis/Inter-subject" folder (top), and three subject averages per subject in the "intra-subject" folders (bottom). 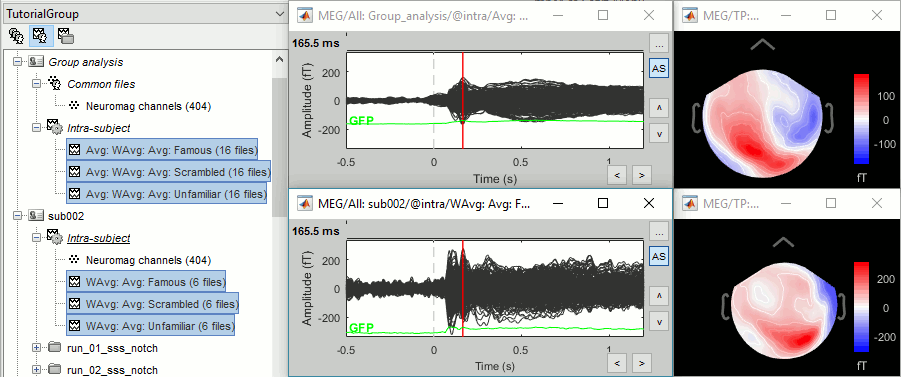
Sources
Keep all the subjects selected in Process1. Select button [Process sources].
If you run the averaging process directly with this file selection, it would average together the MEG and the EEG sources (36 files selected per subject instead of 18). To average separately the two modalities, we can use the Filter box at the bottom-right corner of the Brainstorm window. Enter "EEG" and it would select only the files with a comment including the tag "EEG".
Select process Average > Average files: By trial group (subject average)
EEG source files only, Arithmetic average, Weighted
Add process File > Add tag: EEG.
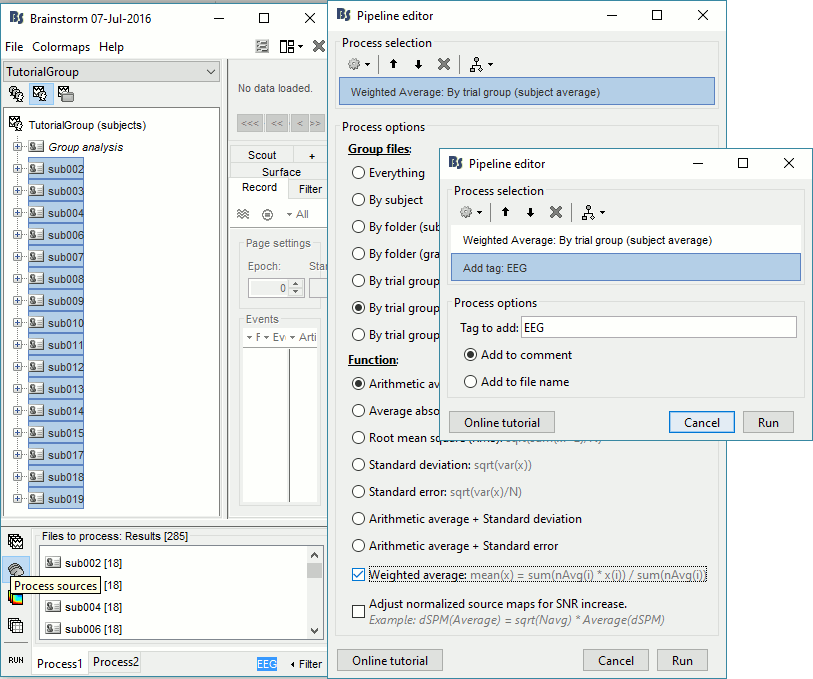
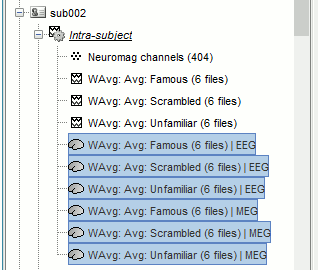
Repeat the same steps with the MEG source files (select all subjects, filter, average, tag).
This time, we cannot compute a grand average directly because the source spaces used to estimate the brain activity do not match between subjects. We have to go through an additional step of projection on a template anatomy. We will also wait to have normalized the source maps before displaying them.
Time-frequency
Keep all the subjects selected in Process1. Select button [Process time-freq].
Make sure you reset the Filter search box at the bottom-right of the Brainstorm window.
Run process Average > Average files: By trial group (subject average)
Arithmetic average, Weighted, Match signals
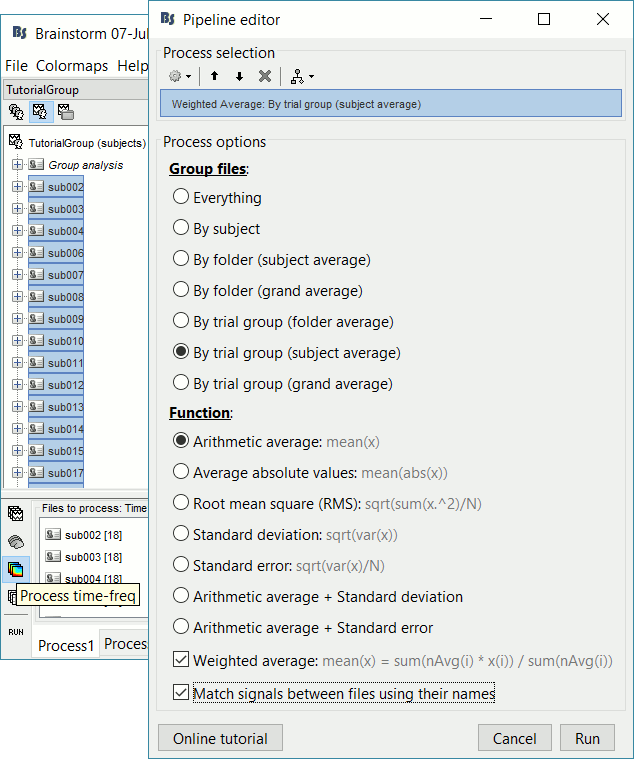
- Like for the sources, we will first normalize the time-frequency maps with respect with a baseline before computing grand averages. Like the sensor data: it is valid to average EEG recordings across subjects, but it is not accurate to average MEG sensors across subjects.
We can average the time-frequency files across runs (it is as valid as averaging the MEG or EEG signals), but we need to select this option "match signals between files using their names" to account for the variable number of bad channels we may have across runs or subjects.
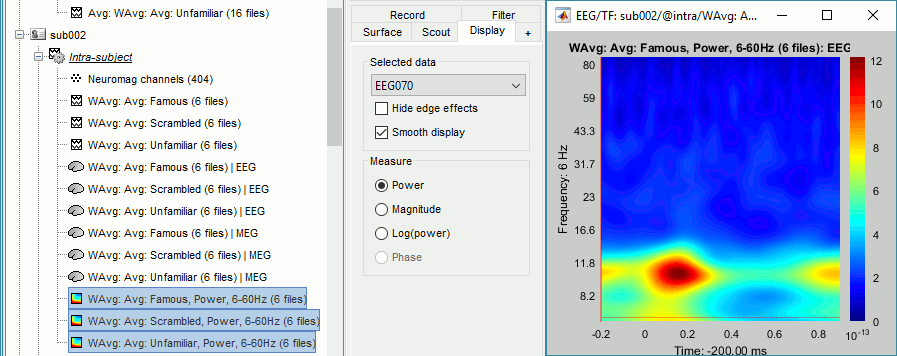
Subject averages: Faces
One of the contrast we want to study is faces (Famous and Unfamiliar) vs non-faces (Scrambled). We need to re-average the Famous and Unfamiliar averages together.
MEG/EEG
- In Process2: FilesA = all the Famous subject averages, FilesB = all the Unfamiliar subject averages. The subjects must be in the same order in the two lists because the process will process the files by pairs of files (A,B).
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces"
Add process Average > Average files: By trial group (subject average): Not weighted
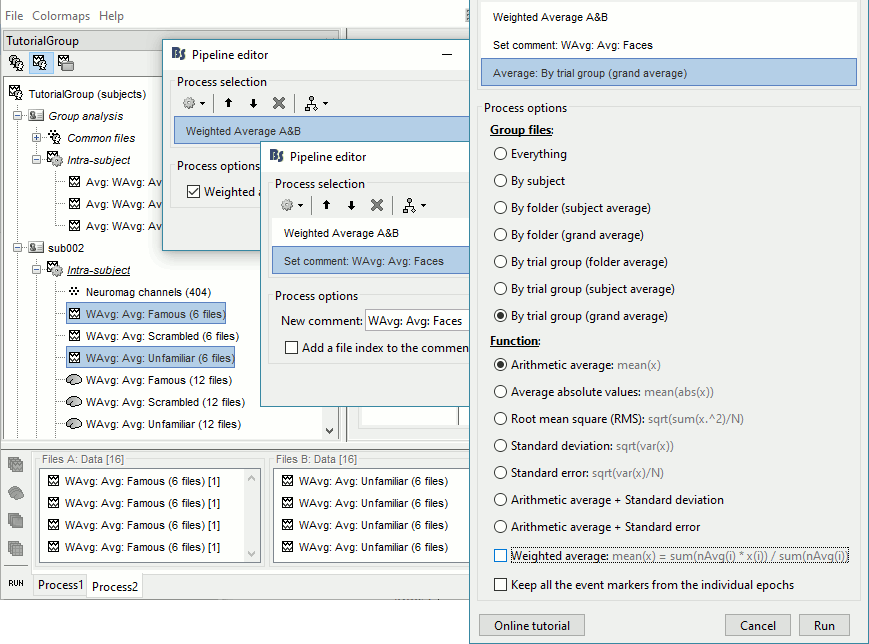
- Selecting that many files from the database explorer can be long when done manually, but can be very efficient from a script. Check out the script equivalent at the end of this page.
It creates one new average for each pair (A,B) and computes the grand average for condition Faces.
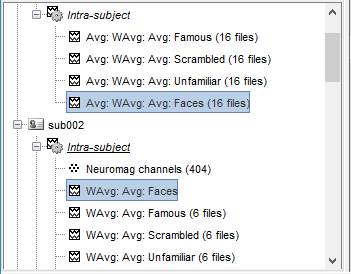
Sources
In Process2: FilesA = all the Famous subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process sources]
- Make sure the order of the files matches in the two lists.
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces | EEG"
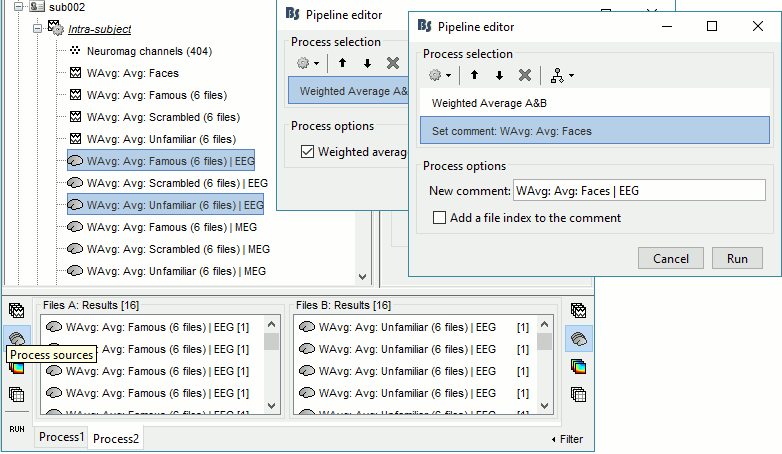
Repeat the same steps for the MEG sources.
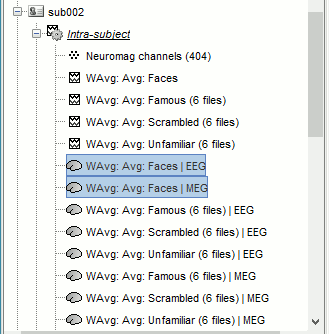
Time-frequency
In Process2: FilesA = all the Famous subject averages (EEG only) [Process timefreq]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process timefreq]
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces"
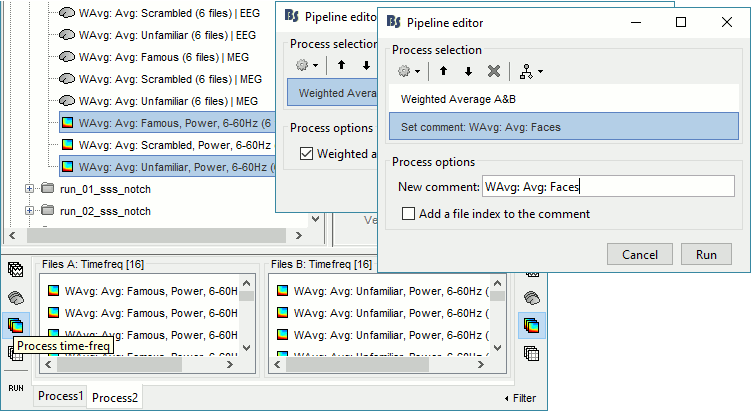
In output: the average for the Face condition, for each subject.

Subject averages: Within-subject differences
To detect correctly the differences between two conditions at the source level, we need to estimate the differences of the conditions for each subject, and then normalize the difference (see tutorial Workflows).
Sources: Faces - Scrambled
In Process2: FilesA = all the Faces subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Scrambled subject averages (EEG only) [Process sources]
Select process Difference > Difference A-B: Do not use absolute values.
Add process File > Set comment: Comment=Faces - Scrambled | EEG
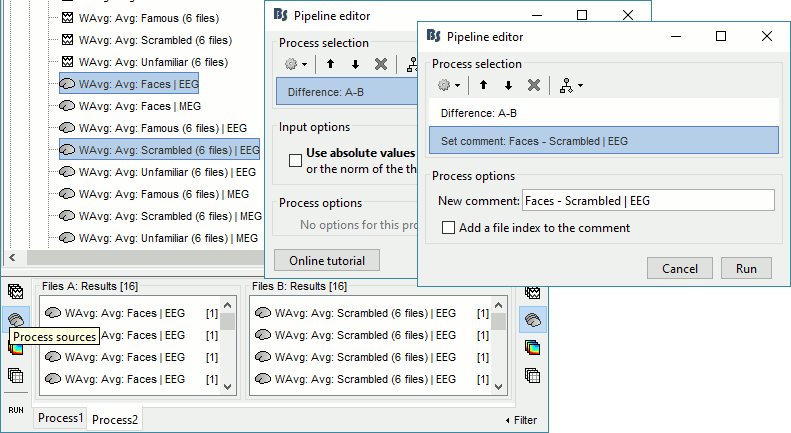
Repeat the same steps for the MEG sources.
Sources: Famous - Unfamiliar
In Process2: FilesA = all the Famous subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process sources]
Select process Difference > Difference A-B: Do not use absolute values.
Add process File > Set comment: Comment=Famous - Unfamiliar | EEG
Repeat the same steps for the MEG sources.
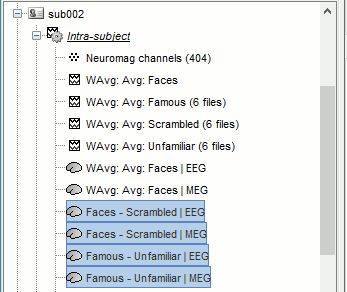
Subject averages: Filter and normalize
Before comparing the averages across subjects we are going to low-pass filter the signals below 32Hz (to smooth possible latency differences between subjects) and normalize the source and time-frequency values with respect with a baseline (see tutorial Workflows).
MEG/EEG
In Process1, select all the Intra-subject folders from all the subjects, select [Process recordings]. For a faster selection, you can use the view "Functional data (sorted by conditions)".
Select process Pre-process > Band-pass filter: 0Hz-32Hz, MEG,EEG, 60dB, No mirror, Overwrite.
Add process Extract > Extract time: Time window=[-200,900]ms
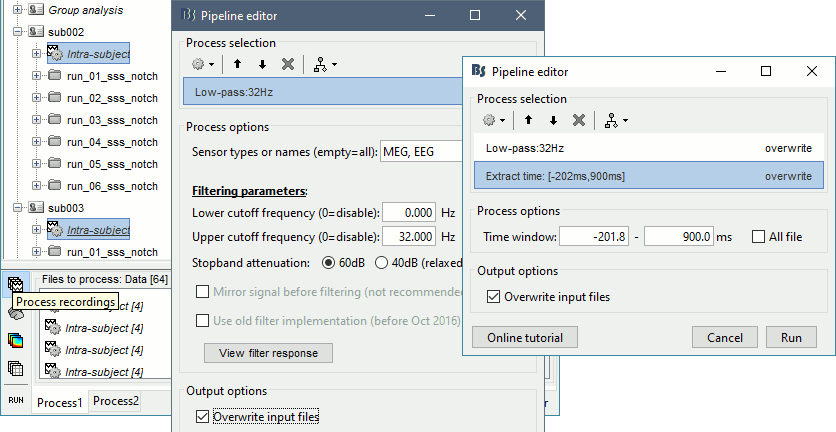
Two tags are added at the end of the comments of the averaged recordings. The selection of a smaller time window gets rid of most of the possible edge effects caused by the filter.
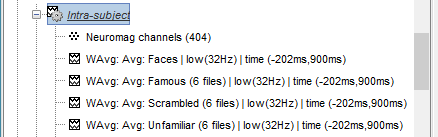
Sources
In Process1, select all the Intra-subject folders from all the subjects, select [Process sources].
Select process Pre-process > Band-pass filter: 0Hz-32Hz, MEG,EEG, Mirror, Overwrite
Add process Extract > Extract time: Time window=[-200,900]ms
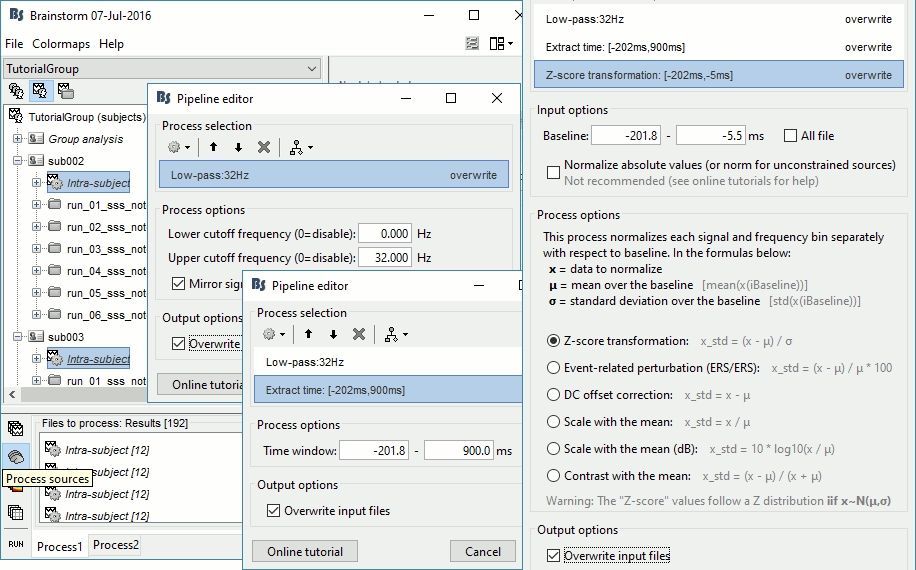
Add process Standardize > Baseline normalization: Baseline=[-200,-5]ms, Z-score, Overwrite
Three tags are added at the end of the comments of the averaged sources.

Time-frequency
In Process1, select all the Intra-subject folders from all the subjects, select [Process time-freq].
Run process Standardize > Baseline normalization: Baseline=[-200,-5]ms, ERS/ERD, Overwrite
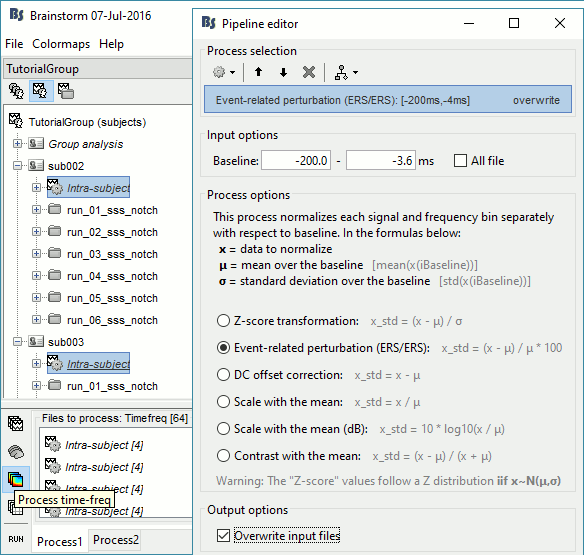
One tag is added at the end of the comments of the averaged time-frequency maps.

Subject averages: Screen captures
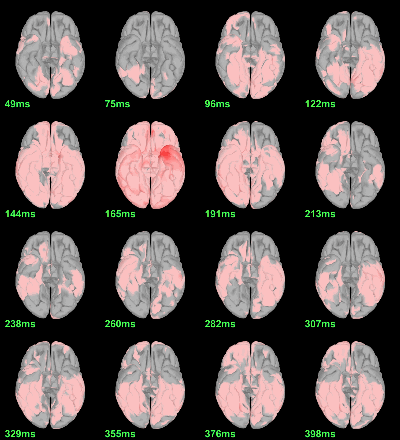
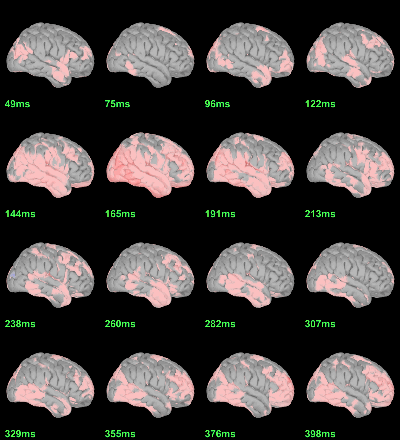
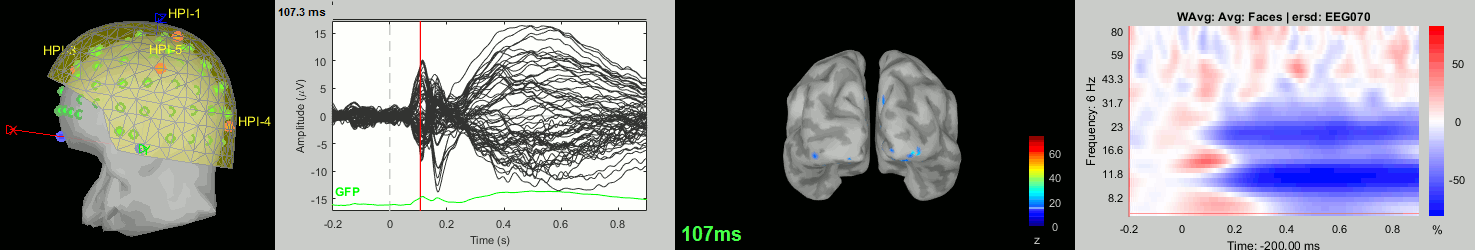
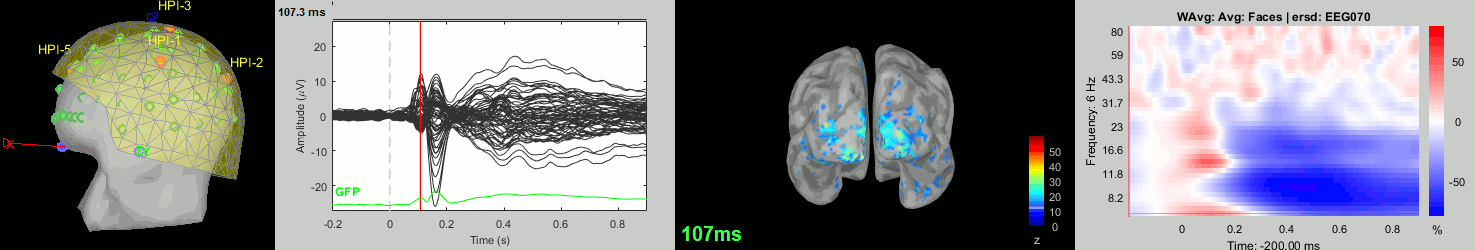
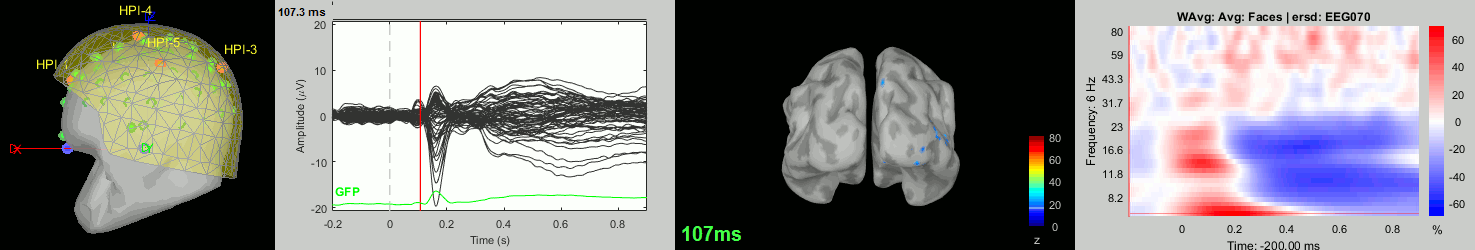
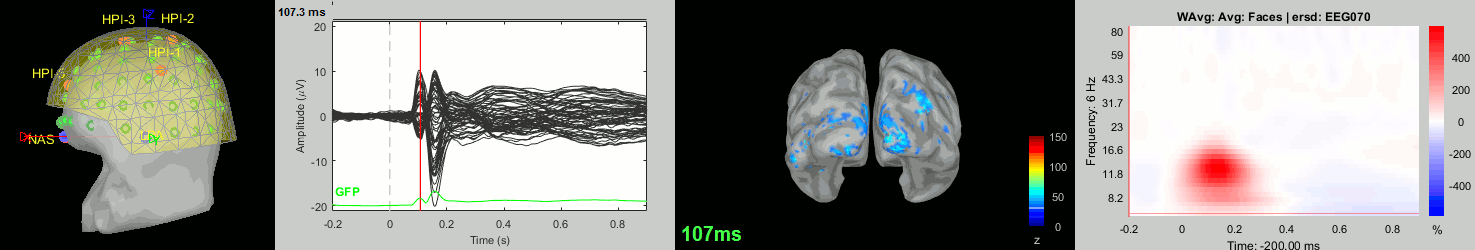
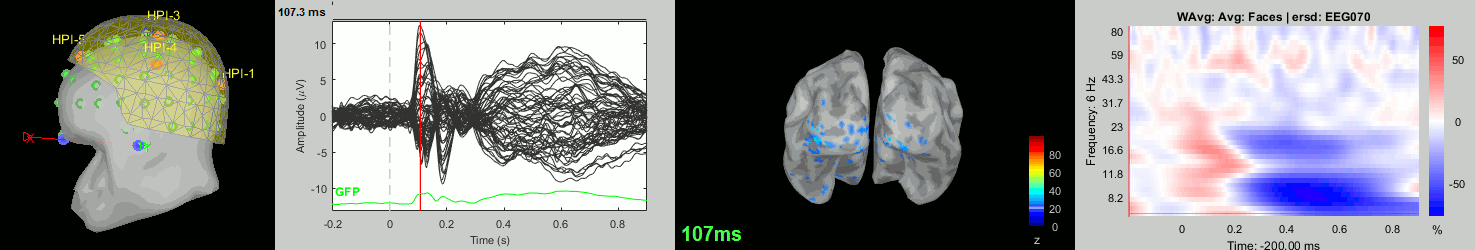
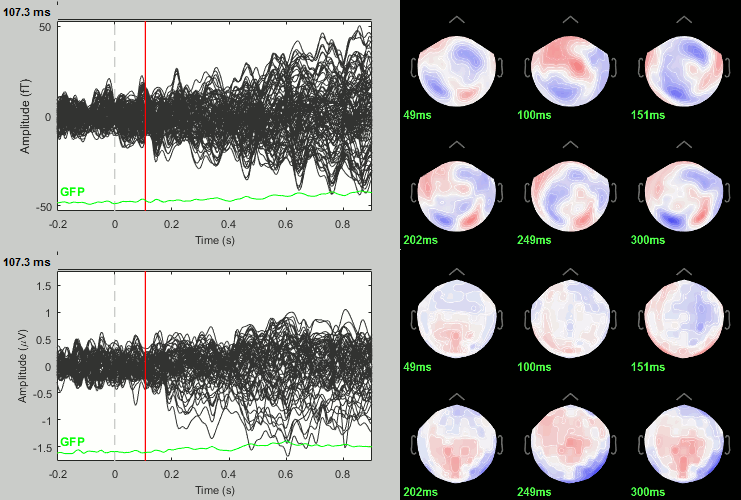
Now we have all the measures ready to be compared across subjects: MEG/EEG, sources, time-frequency. Let's take a few screen captures to make sure the primary visual response look good for all the subjects. The screen captures below represent from left to right:
- Registration sensors/MRI: Head surface, MEG (inside of the helmet), EEG+headpoints (green points).
- Faces: EEG recordings
- Faces: MEG sources at 107ms
- Faces: Time-frequency for EEG070
sub-01
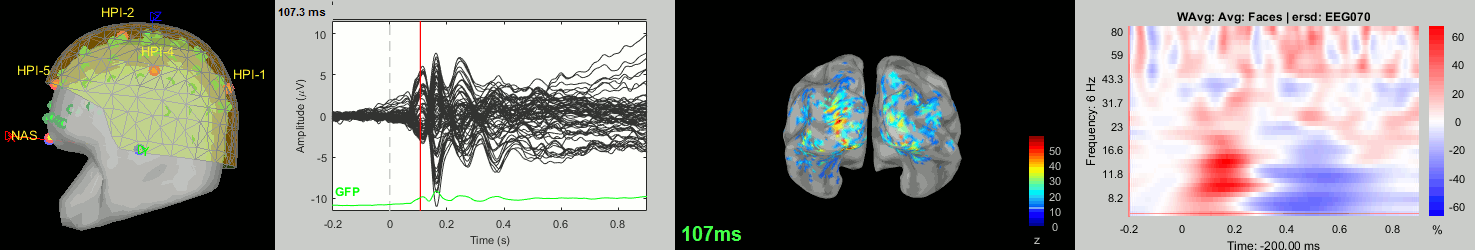
sub-02
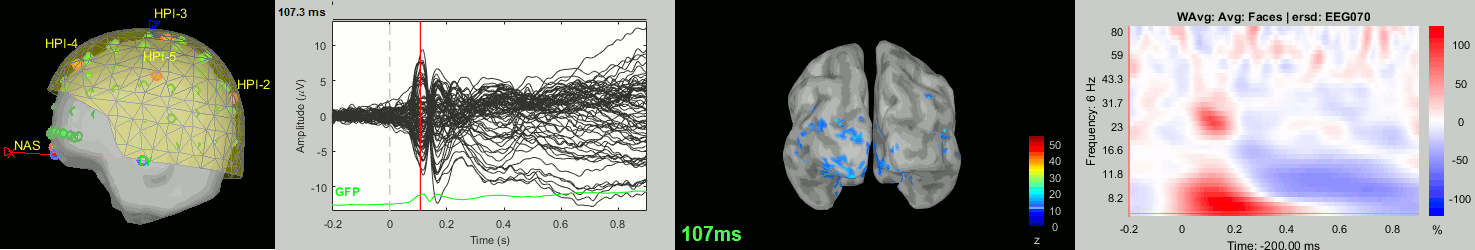
sub-03
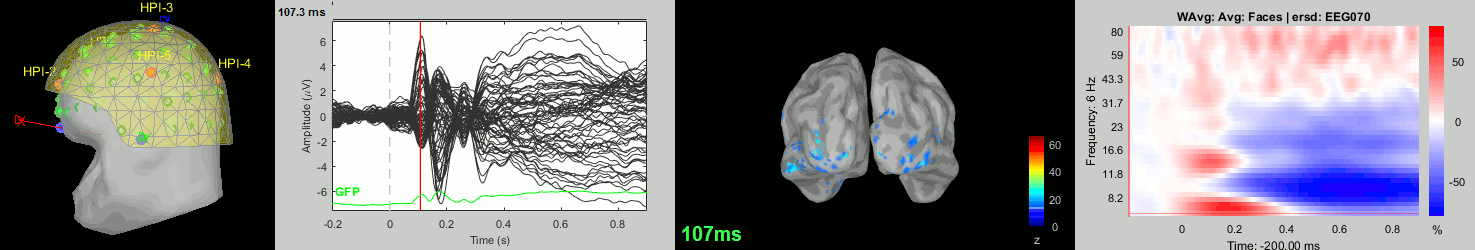
sub-04
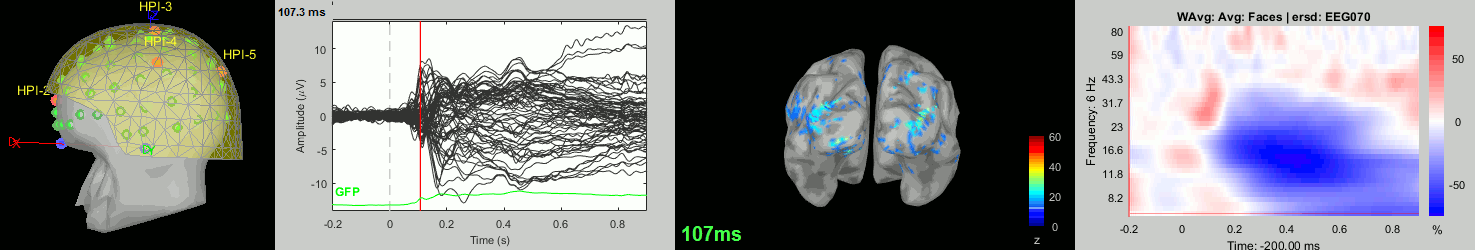
sub-05
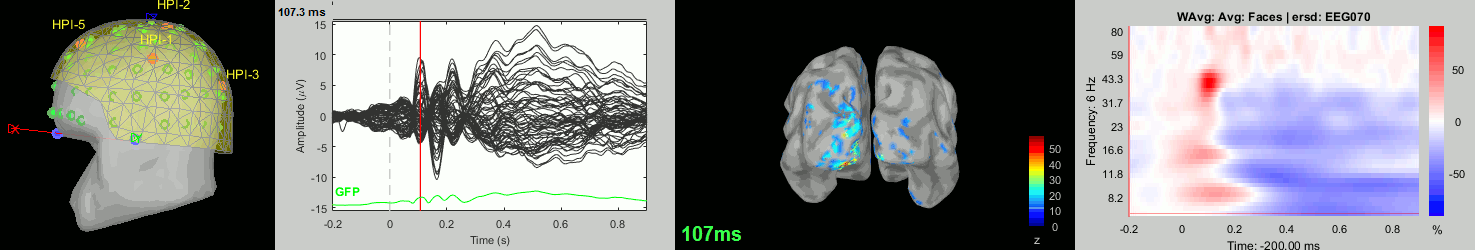
sub-06
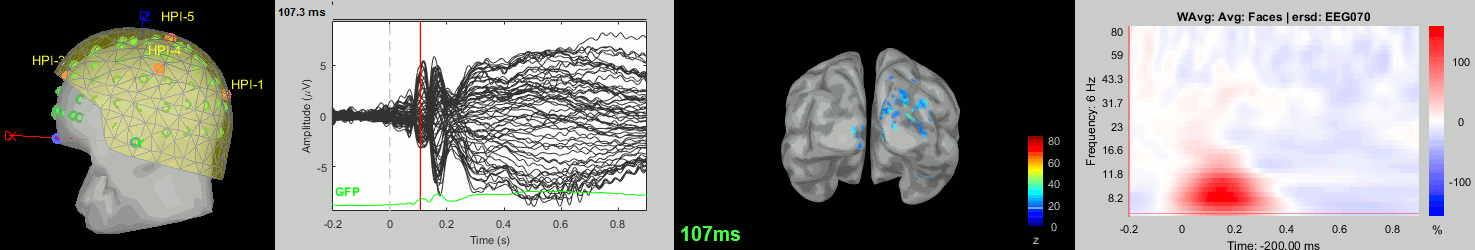
sub-07
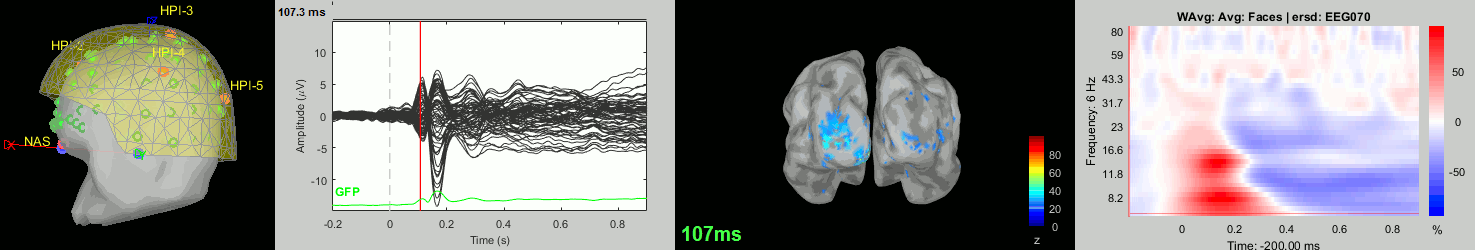
sub-08
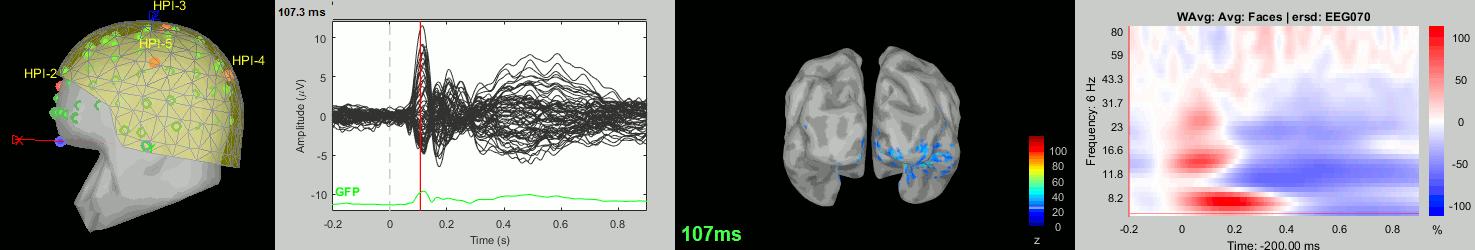
sub-09
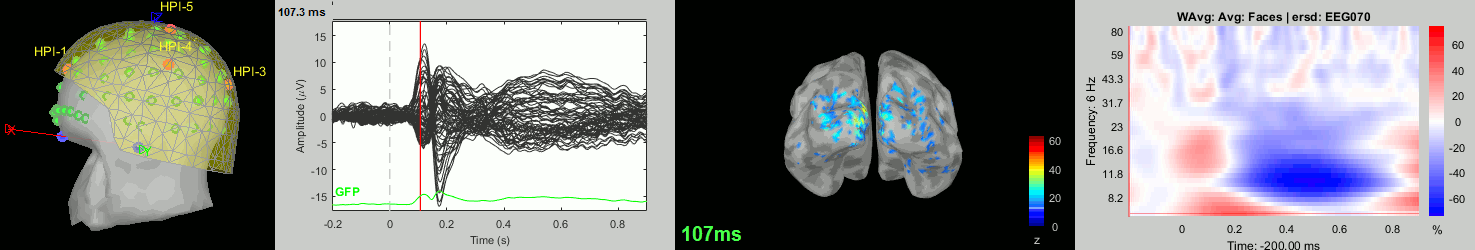
sub-10
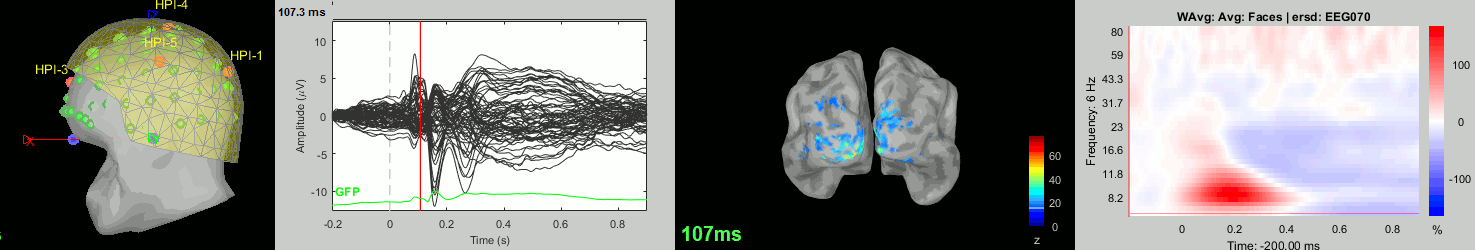
sub-11
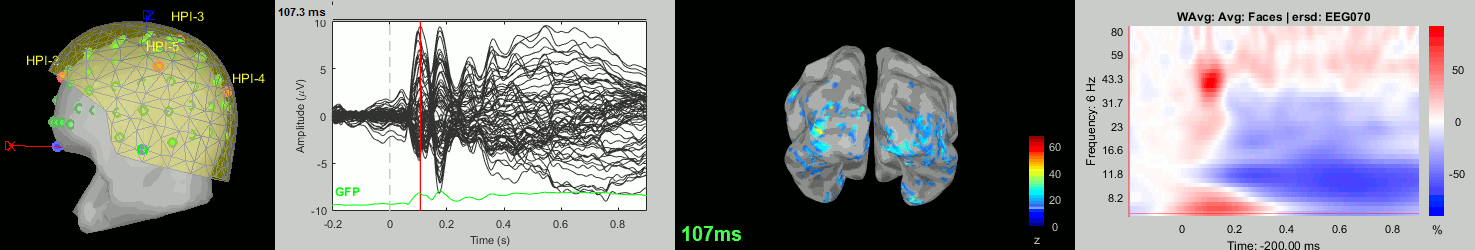
sub-12
sub-13
sub-14
sub-15
sub-16
Group analysis: MEG/EEG
Grand averages
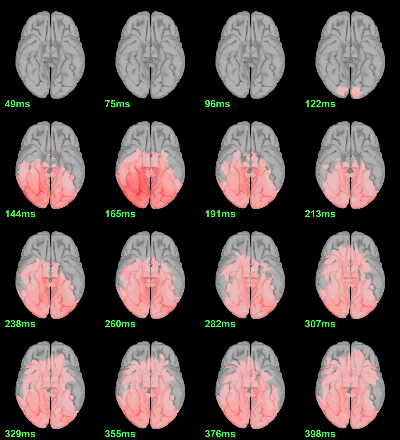
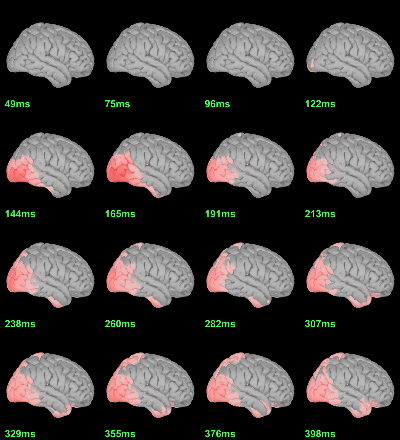
We have already computed the group averages for all the conditions. We will not look much further at these results as we are more interested in the contrasts between conditions. Below are screen captures for all group averages (top=MEG MAG, bottom=EEG). Topography at [50,100,150,200,250,300]ms.
Faces: Famous / Unfamiliar
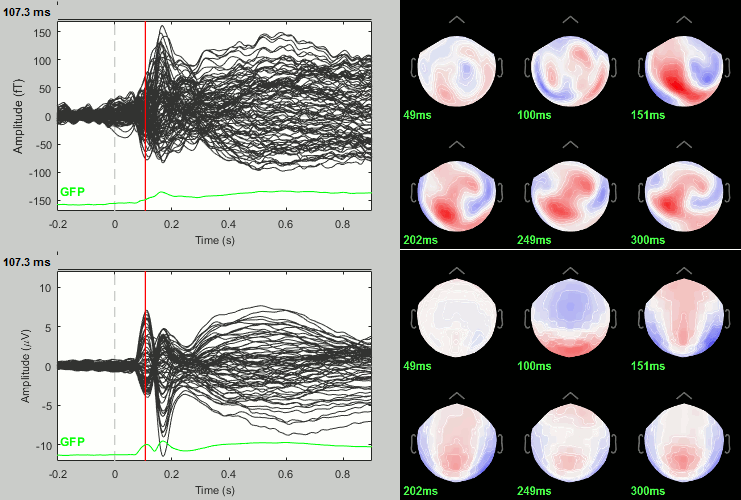
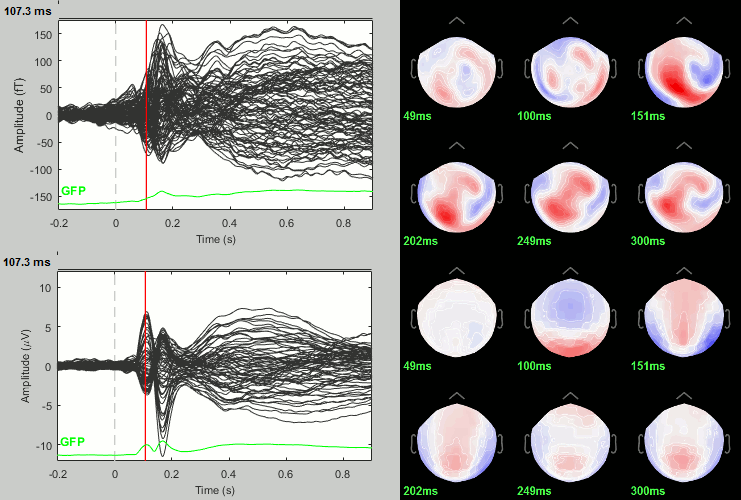
Scrambled
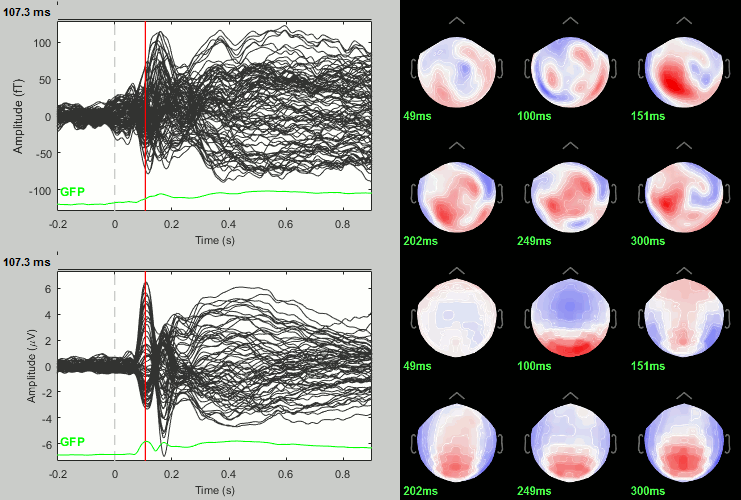
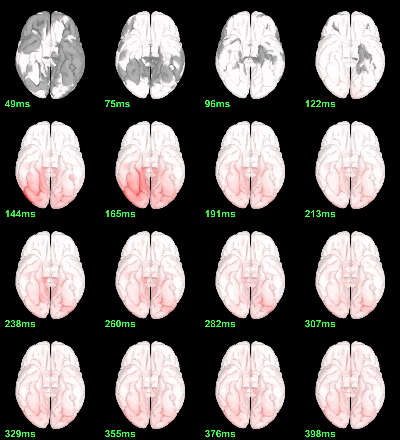
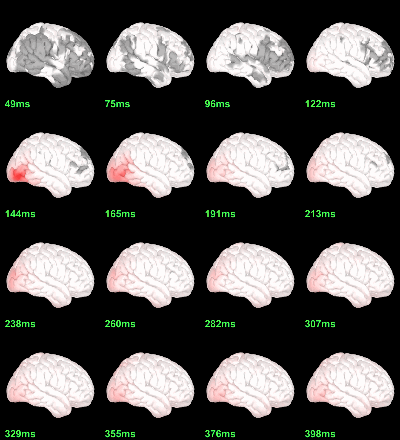
Faces - Scrambled: Differences of averages
We could compute the contrasts directly from the grand averages, but we will do it from the subject averages because it will be the same file selection for the statistics.
- In Process2: FilesA = all the Faces subject averages (from the Intra-subject folders).
- In Process2: FilesB = all the Scrambled subject averages (from the Intra-subject folders).
Run process: Test > Difference of means: Arithmetic average, Not weighted.
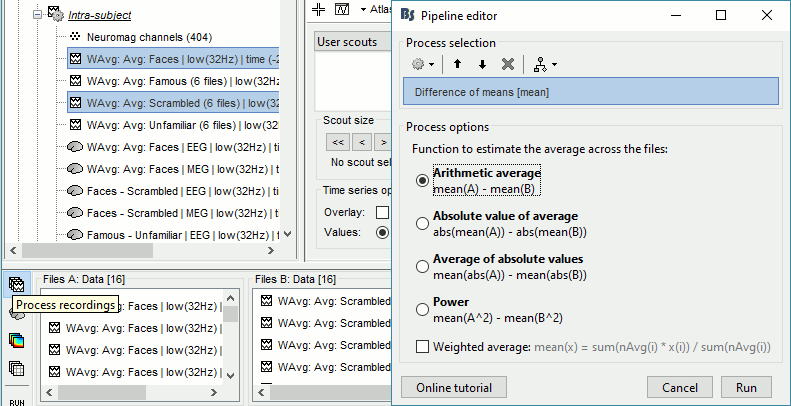
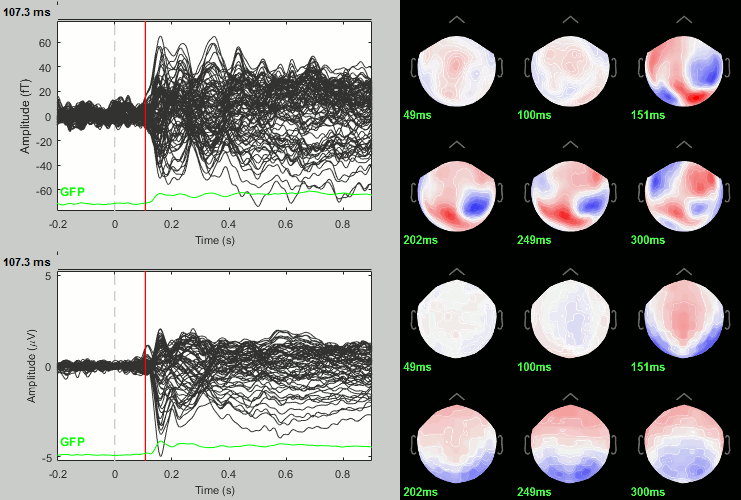
Rename the file: Faces - Scrambled (top=MEG MAG, bottom=EEG).
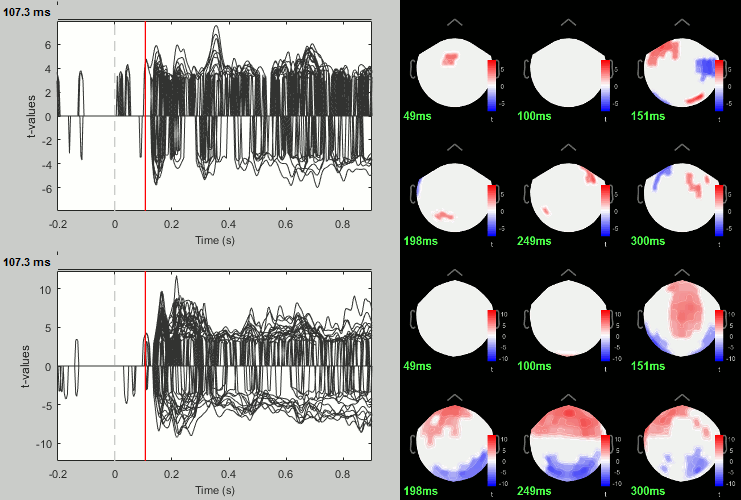
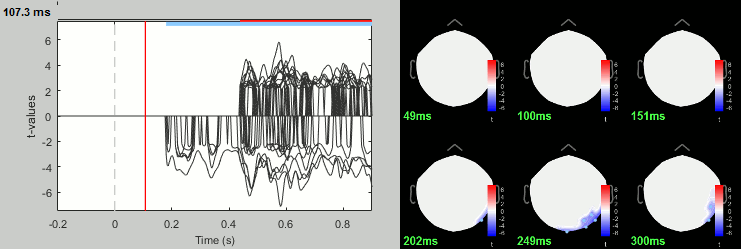
Faces - Scrambled: Significance testing
We have computed the amplitude of the difference between the two conditions, and we will try now to estimate a p-value indicating the significance of each of these values. In all the screen captures below: top=MEG MAG, bottom=EEG.
- In Process2: Keep the same file selection.
Run process: Test > Parametric test: Paired: All file, All sensors, No average, two-tailed.
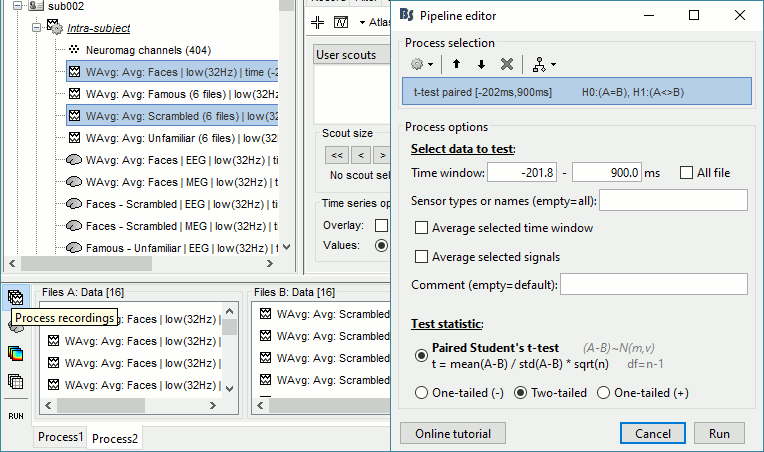
Rename the file: Faces - Scrambled: Parametric t-test. Display with α=0.05, FDR-corrected.
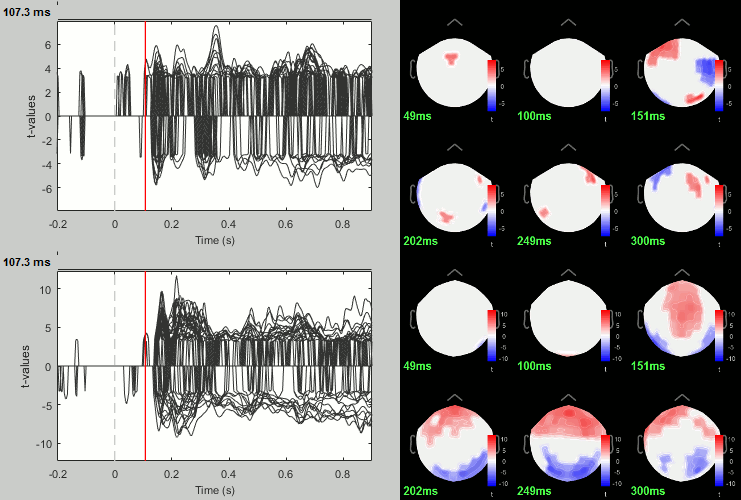
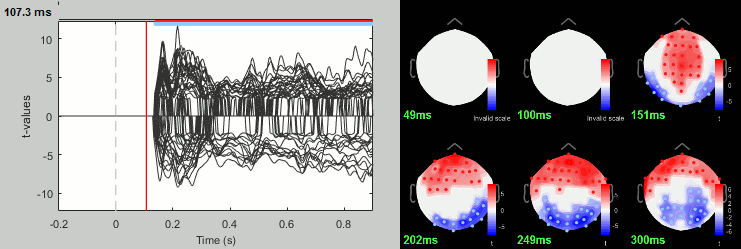
- You can run other tests in a similar way, with almost identical results.
Process: Test > Permutation test: Paired: All file, All sensors, Paired t-test, 1000 randomizations. Display with α=0.05, FDR-corrected.
Process: Test > FieldTrip: ft_timelockstatistics: All file, EEG, Paired t-test, 1000 randomizations, correction=cluster, cluster alpha=0.05. The cluster-based statistics must be executed on one type of sensors at a time (EEG, MEG MAG or MEG GRAD), because it tries to identify spatio-temporal clusters which groups adjacent sensors.
Famous - Unfamiliar: Differences of averages
- In Process2: FilesA = all the Famous subject averages (from the Intra-subject folders).
- In Process2: FilesB = all the Unfamiliar subject averages (from the Intra-subject folders).
Run process: Test > Difference of means: Arithmetic average, Not weighted.
Rename the file: Famous - Unfamiliar.
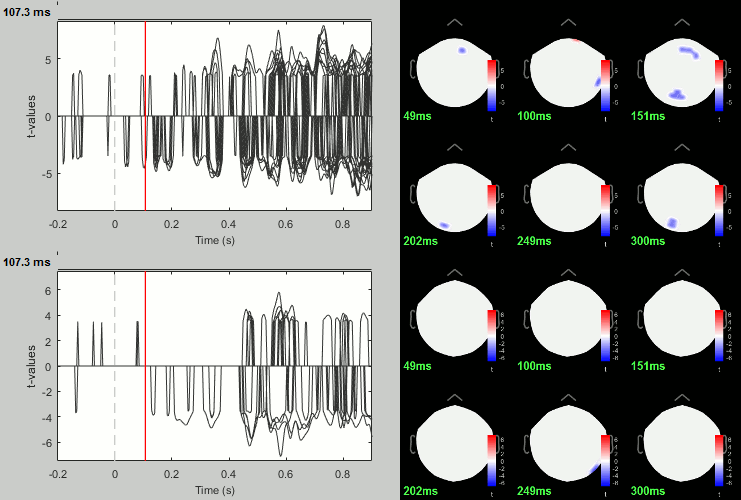
Famous - Unfamiliar: Significance testing
- In Process2: Keep the same file selection.
Run process: Test > Parametric test: Paired: All file, All sensors, No average, two-tailed.
Rename the file: Faces - Scrambled: Parametric t-test. Display with α=0.05, FDR-corrected.
Run process: Test > FieldTrip: ft_timelockstatistics: All file, EEG, Paired t-test, 1000 randomizations, correction=cluster, cluster alpha=0.05.
Group analysis: Sources
Project sources on template
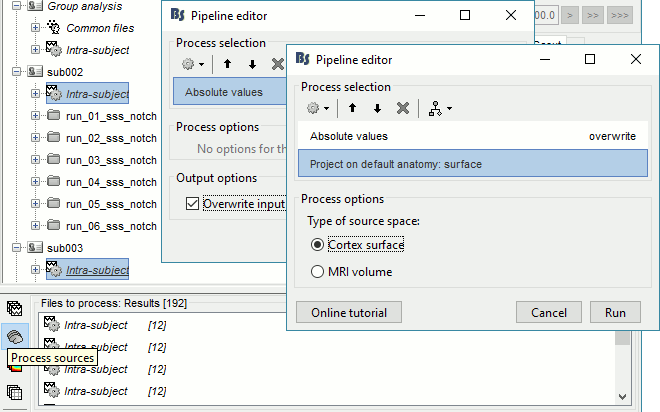
The sources were estimated on the individual anatomy of each subject, the resulting cortical source maps cannot be averaged directly. We need first to re-interpolate all the individual results on a common template (the ICBM152 brain, available in the "default anatomy" folder of the protocol). We also need to extract the absolute values for these source maps: the sign of the minimum norm maps are relative to the orientation of the current with respect to the surface normal, which can vary between subjects.
In Process1, select all the Intra-subject folders from all the subjects, select [Process sources]. For a faster selection, you can use the view "Functional data (sorted by conditions)".
Select process Pre-process > Absolute values: Sensor types=MEG,EEG, Overwrite.
Add process Sources > Project on default anatomy.
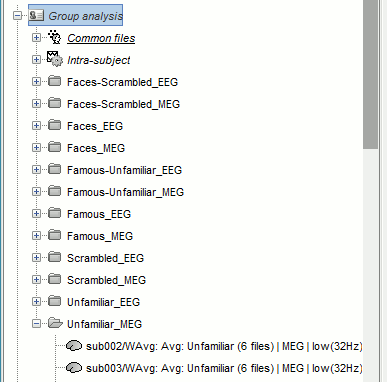
- All the 192 subject averages are projected in the same folder "Group analysis / Intra-subject". Re-organize all the projected source files in new folders, one folder per condition.
To create a folder, right-click on Group analysis > New folder. To move a file to the new folder: drag and drop it in the database explorer or use the keyboard shortcuts Ctrl+X/Ctrl+V.
Spatial smoothing
The source maps estimated with constrained orientations can show very focal activity: two adjacent vertices may have very different normals, and therefore very different current values. When averaging multiple subjects, the peaks of activity may not align very well across subjects. Smoothing spatially the source maps may help obtaining better group results.
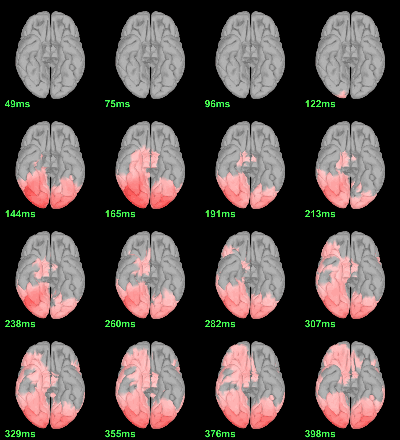
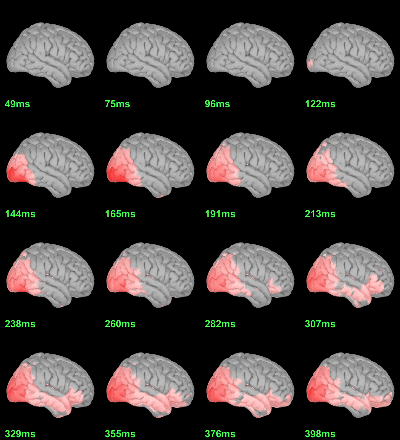
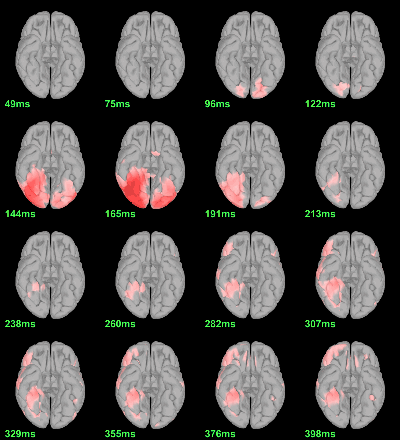
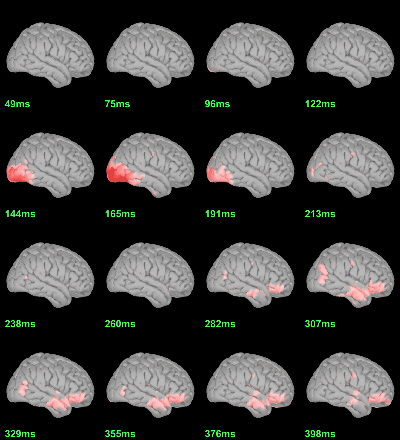
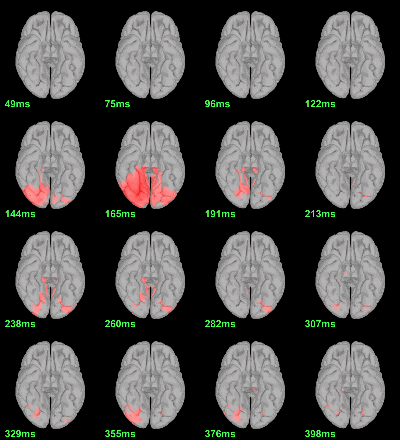
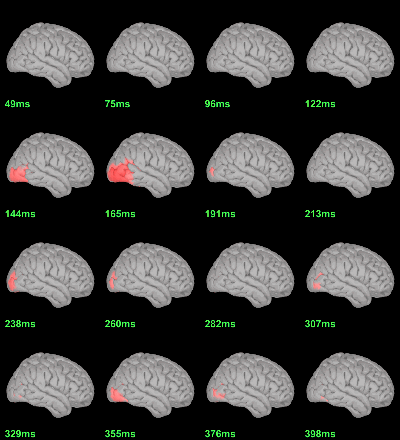
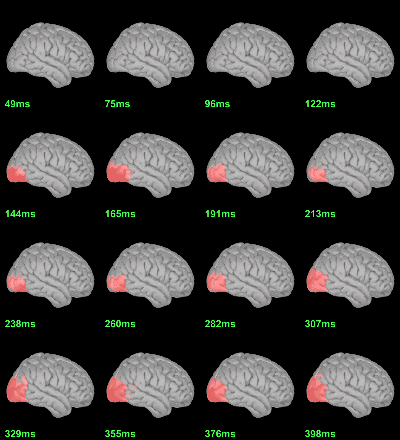
MEG: mean(|Faces-Scrambled|)
MEG: Chi2-test |Faces-Scrambled|=0
- In Process1, select all the source files in Group_analysis/Faces-Scrambled_MEG.
Run process Test > Parametric test against zero: All file, One-sample Chi2-test two-tailed.
Screen capture: α=0.05 FDR-corrected
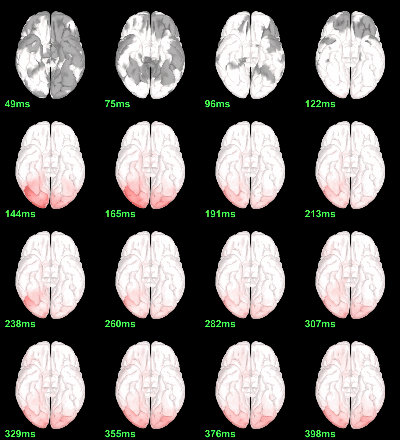
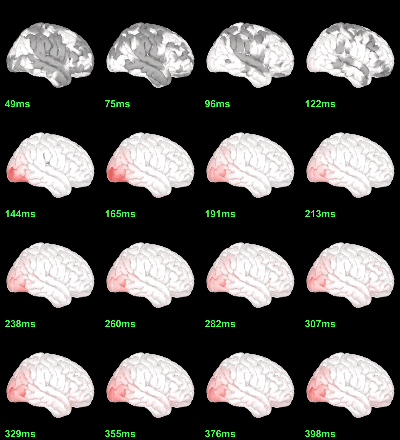
This tests the hypotesis H0:[mean_subject(|Faces-Scrambled|) = 0]
This test is appropriate to detect the differences between the two conditions, but cannot determine which condition is stronger. Its obvious limitation is that it is too sensitive: post-stimulation, everything is shown as significant.
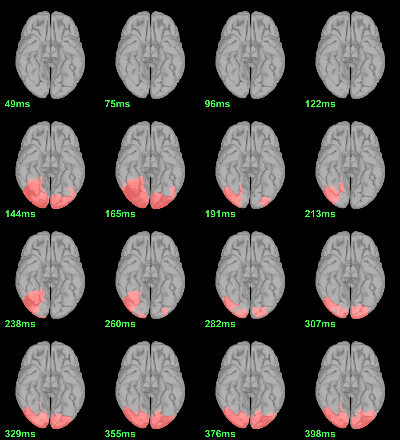
MEG: Chi2-test log(|Faces-Scrambled|)=0
Right-click on the folder Group_analysis/Faces-Scrambled_MEG > File > Duplicate folder.
Rename the new folder: Faces-Scrambled_MEG_log.
- In Process1, select all the source files in Group_analysis/Faces-Scrambled_MEG_log.
Select process Pre-process > Run Matlab command > "Data = log(Data)";
Add process Test > Parametric test against zero: All file, One-sample Chi2-test two-tailed.
Screen capture: α=0.05, FDR-corrected
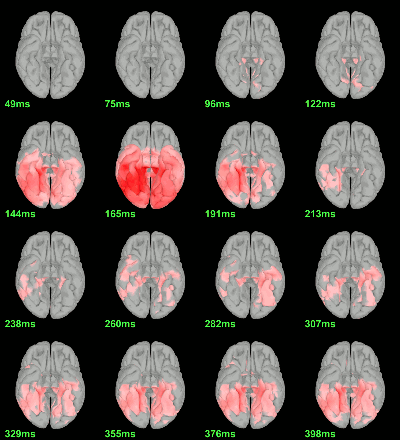
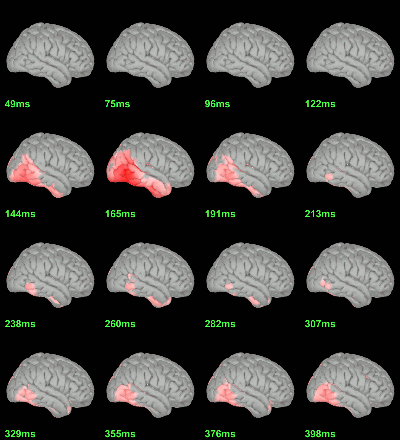
MEG: mean(|Faces|)-mean(|Scrambled|)
MEG: Student t-test |Faces|=|Scrambled|
- In Process2: FilesA = all the source files in Group_analysis/Faces_MEG.
- In Process2: FilesB = all the source files in Group_analysis/Scrambled_MEG.
Run process Test > Parametric test: Paired: All file, No average, two-tailed.
Screen capture: α=0.05 FDR-corrected
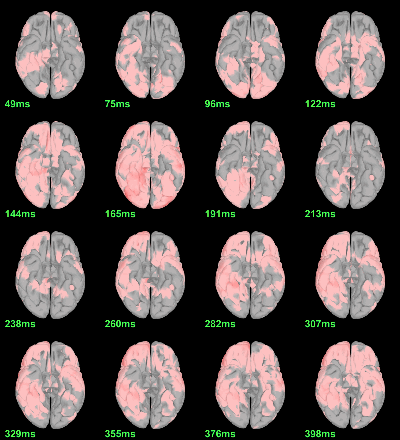
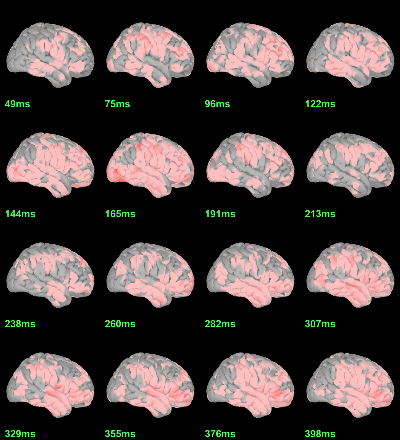
This test the hypothesis H0:[mean(abs(Faces))=mean(abs(Scrambled))].
This measure is not appropriate to properly identify the differences between the two conditions because it is insensitive to the sign of the sources within each subject.
EEG: Faces-Scrambled
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_visual_group.m
Execution reports: Subject averages, MEG/EEG group results, Source group results
1 function tutorial_visual_group(ProtocolName, reports_dir)
2 % TUTORIAL_VISUAL_GROUP: Runs the Brainstorm/SPM group analysis pipeline (group analysis, BIDS VERSION)
3 %
4 % ONLINE TUTORIALS:
5 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle
6 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup
7 %
8 % INPUTS:
9 % - ProtocolName : Name of the protocol in which the recordings for the 19 subjects have been imported (TutorialVisual or TutorialGroup)
10 % This folder must include: the run-level averages (recordings and time-frequency) and the source kernels for each run
11 % - reports_dir : If defined, exports all the reports as HTML to this folder
12
13 % @=============================================================================
14 % This function is part of the Brainstorm software:
15 % https://neuroimage.usc.edu/brainstorm
16 %
17 % Copyright (c) University of Southern California & McGill University
18 % This software is distributed under the terms of the GNU General Public License
19 % as published by the Free Software Foundation. Further details on the GPLv3
20 % license can be found at http://www.gnu.org/copyleft/gpl.html.
21 %
22 % FOR RESEARCH PURPOSES ONLY. THE SOFTWARE IS PROVIDED "AS IS," AND THE
23 % UNIVERSITY OF SOUTHERN CALIFORNIA AND ITS COLLABORATORS DO NOT MAKE ANY
24 % WARRANTY, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO WARRANTIES OF
25 % MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE, NOR DO THEY ASSUME ANY
26 % LIABILITY OR RESPONSIBILITY FOR THE USE OF THIS SOFTWARE.
27 %
28 % For more information type "brainstorm license" at command prompt.
29 % =============================================================================@
30 %
31 % Author: Francois Tadel, Elizabeth Bock, 2016-2018
32
33 % ===== CHECK PROTOCOL =====
34 % Start Brainstorm without the GUI
35 if ~brainstorm('status')
36 brainstorm nogui
37 end
38 % Output folder for reports
39 if (nargin < 2) || isempty(reports_dir) || ~isdir(reports_dir)
40 reports_dir = [];
41 end
42 % You have to specify the folder in which the tutorial dataset is unzipped
43 if (nargin < 1) || isempty(ProtocolName)
44 ProtocolName = 'TutorialGroup';
45 end
46 % Select current protocol
47 iProtocol = bst_get('Protocol', ProtocolName);
48 if isempty(iProtocol)
49 error(['Unknown protocol: ' ProtocolName]);
50 end
51 if (iProtocol ~= bst_get('iProtocol'))
52 gui_brainstorm('SetCurrentProtocol', iProtocol);
53 end
54 % Process: Select results files in: sub-01/sub-01_ses-meg_task-facerecognition_run-01_proc-sss_meg_notch
55 sFiles = bst_process('CallProcess', 'process_select_files_results', [], [], ...
56 'subjectname', 'sub-01', ...
57 'condition', 'sub-01_ses-meg_task-facerecognition_run-01_proc-sss_meg_notch');
58 if isempty(sFiles)
59 error(['No source files available in folder: sub-01/sub-01_ses-meg_task-facerecognition_run-01_proc-sss_meg_notch.' 10 ...
60 'You should run tutorial_visual_single first, or download the ' 10 ...
61 'protocol TutorialGroup.zip from the Brainstorm website.']);
62 end
63 % Process: Select file comments with tag: Unconstr
64 sFiles = bst_process('CallProcess', 'process_select_tag', sFiles, [], 'tag', 'Unconstr');
65 if ~isempty(sFiles)
66 isUnconstrained = 1;
67 else
68 isUnconstrained = 0;
69 end
70 % Start a new report
71 bst_report('Start');
72
73
74 % ===== DELETE ALL THE EXISTING FILES =====
75 GroupSubjectName = bst_get('NormalizedSubjectName');
76 DirIntra = bst_get('DirAnalysisIntra');
77 % Delete group analysis
78 [sSubject, iSubject] = bst_get('Subject', GroupSubjectName);
79 if ~isempty(sSubject)
80 db_delete_subjects(iSubject);
81 end
82 % Get all the files in the intra subjects
83 sAvgSrc = bst_process('CallProcess', 'process_select_files_data', [], [], ...
84 'subjectname', 'All', ...
85 'condition', DirIntra, ...
86 'includeintra', 1);
87 % If there are existing files: delete the contents of the folders
88 if ~isempty(sAvgSrc)
89 % Get the base folder of the protocol
90 ProtocolInfo = bst_get('ProtocolInfo', iProtocol);
91 % Get all the folders
92 AllIntraDir = unique(cellfun(@bst_fileparts, {sAvgSrc.FileName}, 'UniformOutput', 0));
93 % Loop over the intra folders
94 for iSubj = 1:length(AllIntraDir)
95 % Get all the files
96 subjIntraDir = dir(fullfile(ProtocolInfo.STUDIES, AllIntraDir{iSubj}, '*.mat'));
97 subjIntraFiles = setdiff({subjIntraDir.name}, 'brainstormstudy.mat');
98 % Delete all the files
99 for iFile = 1:length(subjIntraFiles)
100 delete(fullfile(ProtocolInfo.STUDIES, AllIntraDir{iSubj}, subjIntraFiles{iFile}));
101 end
102 end
103 % Reload protocol
104 db_reload_database(iProtocol);
105 end
106
107
108 %% ===== SUBJECT AVERAGES: FAMOUS, UNFAMILIAR, SCRAMBLED ==============================
109 % ====================================================================================
110
111 % ===== SUBJECT AVERAGE: MEG+EEG =====
112 % Process: Select data files in: */*/Avg
113 sAvgRun = bst_process('CallProcess', 'process_select_files_data', [], [], ...
114 'subjectname', 'All', ...
115 'tag', 'Avg');
116 % Process: Weighted Average: By trial group (subject average)
117 sAvgSubj = bst_process('CallProcess', 'process_average', sAvgRun, [], ...
118 'avgtype', 6, ... % By trial group (subject average)
119 'avg_func', 1, ... % Arithmetic average: mean(x)
120 'weighted', 1, ...
121 'keepevents', 1);
122 % Process: Average: By trial group (grand average)
123 sAvgGroup = bst_process('CallProcess', 'process_average', sAvgSubj, [], ...
124 'avgtype', 7, ... % By trial group (grand average)
125 'avg_func', 1, ... % Arithmetic average: mean(x)
126 'weighted', 0, ...
127 'keepevents', 0);
128
129 % ===== SUBJECT AVERAGES: SOURCES (EEG) =====
130 % Process: Select source files in: */*/EEG
131 sAvgRunSrcEeg = bst_process('CallProcess', 'process_select_files_results', [], [], 'tag', 'EEG');
132 % Process: Weighted Average: By trial group (subject average) - EEG
133 sAvgSubjSrcEeg = bst_process('CallProcess', 'process_average', sAvgRunSrcEeg, [], ...
134 'avgtype', 6, ... % By trial group (subject average)
135 'avg_func', 1, ... % Arithmetic average: mean(x)
136 'weighted', 1, ...
137 'scalenormalized', 0);
138 % Process: Add tag: EEG
139 sAvgSubjSrcEeg = bst_process('CallProcess', 'process_add_tag', sAvgSubjSrcEeg, [], ...
140 'tag', 'EEG', ...
141 'output', 1); % Add to comment
142
143 % ===== SUBJECT AVERAGES: SOURCES (MEG) =====
144 % Process: Select source files in: */*/MEG
145 sAvgRunSrcMeg = bst_process('CallProcess', 'process_select_files_results', [], [], 'tag', 'MEG');
146 % Process: Weighted Average: By trial group (subject average) - MEG
147 sAvgSubjSrcMeg = bst_process('CallProcess', 'process_average', sAvgRunSrcMeg, [], ...
148 'avgtype', 6, ... % By trial group (subject average)
149 'avg_func', 1, ... % Arithmetic average: mean(x)
150 'weighted', 1, ...
151 'scalenormalized', 1);
152 % Process: Add tag: MEG
153 sAvgSubjSrcMeg = bst_process('CallProcess', 'process_add_tag', sAvgSubjSrcMeg, [], ...
154 'tag', 'MEG', ...
155 'output', 1); % Add to comment
156
157 % ===== SUBJECT AVERAGES: TIMEFREQ =====
158 % Process: Select time-frequency files in: */*
159 sAvgRunTf = bst_process('CallProcess', 'process_select_files_timefreq', [], []);
160 % Process: Weighted Average: By trial group (subject average)
161 sAvgSubjTf = bst_process('CallProcess', 'process_average', sAvgRunTf, [], ...
162 'avgtype', 6, ... % By trial group (subject average)
163 'avg_func', 1, ... % Arithmetic average: mean(x)
164 'weighted', 1, ...
165 'scalenormalized', 1);
166
167
168
169 %% ===== SUBJECT AVERAGES: FACES ======================================================
170 % ====================================================================================
171
172 % ===== FACES AVERAGE: MEG+EEG ======
173 % Process: Select data files in: */*
174 sAllData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
175 'subjectname', 'All', ...
176 'includeintra', 1);
177 % Process: Select file comments with tag: "WAvg: Avg: Famous (6 files)"
178 sAvgSubjFamous = bst_process('CallProcess', 'process_select_tag', sAllData, [], 'tag', 'WAvg: Avg: Famous (6 files)');
179 % Process: Select file comments with tag: "WAvg: Avg: Unfamiliar (6 files)"
180 sAvgSubjUnfamiliar = bst_process('CallProcess', 'process_select_tag', sAllData, [], 'tag', 'WAvg: Avg: Unfamiliar (6 files)');
181 % Process: Weighted Average A&B
182 sAvgSubjFaces = bst_process('CallProcess', 'process_average_ab', sAvgSubjFamous, sAvgSubjUnfamiliar, ...
183 'weighted', 1);
184 % Process: Add tag: Faces
185 sAvgSubjFaces = bst_process('CallProcess', 'process_add_tag', sAvgSubjFaces, [], ...
186 'tag', 'Faces', ...
187 'output', 2); % Add to file name
188 % Process: Set comment
189 sAvgSubjFaces = bst_process('CallProcess', 'process_set_comment', sAvgSubjFaces, [], ...
190 'tag', 'WAvg: Avg: Faces', ...
191 'isindex', 0);
192 % Process: Average: By trial group (grand average)
193 sAvgGroupFaces = bst_process('CallProcess', 'process_average', sAvgSubjFaces, [], ...
194 'avgtype', 7, ... % By trial group (grand average)
195 'avg_func', 1, ... % Arithmetic average: mean(x)
196 'weighted', 0, ...
197 'keepevents', 0);
198
199 % ===== FACES AVERAGE: SOURCES (EEG) ======
200 % Process: Select source files in: */*/EEG
201 sSrcEeg = bst_process('CallProcess', 'process_select_files_results', [], [], ...
202 'subjectname', 'All', ...
203 'tag', 'EEG', ...
204 'includeintra', 1);
205 % Process: Select file comments with tag: "WAvg: Avg: Famous"
206 sAvgSubjFamousSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Famous');
207 % Process: Select file comments with tag: "WAvg: Avg: Unfamiliar"
208 sAvgSubjUnfamiliarSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Unfamiliar');
209 % Process: Weighted Average A&B
210 sAvgSubjFacesSrcEeg = bst_process('CallProcess', 'process_average_ab', sAvgSubjFamousSrcEeg, sAvgSubjUnfamiliarSrcEeg, ...
211 'weighted', 1, ...
212 'scalenormalized', 1);
213 % Process: Add tag: Faces
214 sAvgSubjFacesSrcEeg = bst_process('CallProcess', 'process_add_tag', sAvgSubjFacesSrcEeg, [], ...
215 'tag', 'Faces', ...
216 'output', 2); % Add to file name
217 % Process: Set comment
218 sAvgSubjFacesSrcEeg = bst_process('CallProcess', 'process_set_comment', sAvgSubjFacesSrcEeg, [], ...
219 'tag', 'WAvg: Avg: Faces | EEG', ...
220 'isindex', 0);
221
222 % ===== FACES AVERAGE: SOURCES (MEG) ======
223 % Process: Select source files in: */*
224 sSrcMeg = bst_process('CallProcess', 'process_select_files_results', [], [], ...
225 'subjectname', 'All', ...
226 'tag', 'MEG', ...
227 'includeintra', 1);
228 % Process: Select file comments with tag: Famous
229 sAvgSubjFamousSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'Famous');
230 % Process: Select file comments with tag: Unfamiliar
231 sAvgSubjUnfamiliarSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'Unfamiliar');
232 % Process: Weighted Average A&B
233 sAvgSubjFacesSrcMeg = bst_process('CallProcess', 'process_average_ab', sAvgSubjFamousSrcMeg, sAvgSubjUnfamiliarSrcMeg, ...
234 'weighted', 1, ...
235 'scalenormalized', 1);
236 % Process: Add tag: Faces
237 sAvgSubjFacesSrcMeg = bst_process('CallProcess', 'process_add_tag', sAvgSubjFacesSrcMeg, [], ...
238 'tag', 'Faces', ...
239 'output', 2); % Add to file name
240 % Process: Set comment
241 sAvgSubjFacesSrcMeg = bst_process('CallProcess', 'process_set_comment', sAvgSubjFacesSrcMeg, [], ...
242 'tag', 'WAvg: Avg: Faces | MEG', ...
243 'isindex', 0);
244
245 % ===== FACES AVERAGE: TIMEFREQ ======
246 % Process: Select time-frequency files in: */*
247 sAllTf = bst_process('CallProcess', 'process_select_files_timefreq', [], [], ...
248 'subjectname', 'All', ...
249 'includeintra', 1);
250 % Process: Select file comments with tag: "WAvg: Avg: Famous"
251 sAvgSubjFamousTf = bst_process('CallProcess', 'process_select_tag', sAllTf, [], 'tag', 'WAvg: Avg: Famous');
252 % Process: Select file comments with tag: "WAvg: Avg: Unfamiliar"
253 sAvgSubjUnfamiliarTf = bst_process('CallProcess', 'process_select_tag', sAllTf, [], 'tag', 'WAvg: Avg: Unfamiliar');
254 % Process: Weighted Average A&B
255 sAvgSubjFacesTf = bst_process('CallProcess', 'process_average_ab', sAvgSubjFamousTf, sAvgSubjUnfamiliarTf, ...
256 'weighted', 1);
257 % Process: Add tag: Faces
258 sAvgSubjFacesTf = bst_process('CallProcess', 'process_add_tag', sAvgSubjFacesTf, [], ...
259 'tag', 'Faces', ...
260 'output', 2); % Add to file name
261 % Process: Set comment
262 sAvgSubjFacesTf = bst_process('CallProcess', 'process_set_comment', sAvgSubjFacesTf, [], ...
263 'tag', 'WAvg: Avg: Faces', ...
264 'isindex', 0);
265
266
267
268 %% ===== SUBJECT AVERAGES: WITHIN-SUBJECT DIFFERENCES =================================
269 % ====================================================================================
270
271 % ===== FACES - SCRAMBLED: SOURCES EEG =====
272 % Process: Select source files in: */*/EEG
273 sSrcEeg = bst_process('CallProcess', 'process_select_files_results', [], [], ...
274 'subjectname', 'All', ...
275 'tag', 'EEG', ...
276 'includeintra', 1);
277 % Process: Select file comments with tag: "WAvg: Avg: Faces"
278 sAvgSubjFacesSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Faces');
279 % Process: Select file comments with tag: "WAvg: Avg: Scrambled"
280 sAvgSubjScrambledSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Scrambled');
281 % Process: Difference: A-B
282 sDiffFacesSubjSrcEeg = bst_process('CallProcess', 'process_diff_ab', sAvgSubjFacesSrcEeg, sAvgSubjScrambledSrcEeg, ...
283 'source_abs', 0);
284 % Process: Set comment
285 sDiffFacesSubjSrcEeg = bst_process('CallProcess', 'process_set_comment', sDiffFacesSubjSrcEeg, [], ...
286 'tag', 'Faces - Scrambled | EEG', ...
287 'isindex', 0);
288
289 % ===== FACES - SCRAMBLED: SOURCES MEG =====
290 % Process: Select source files in: */*/MEG
291 sSrcMeg = bst_process('CallProcess', 'process_select_files_results', [], [], ...
292 'subjectname', 'All', ...
293 'tag', 'MEG', ...
294 'includeintra', 1);
295 % Process: Select file comments with tag: "WAvg: Avg: Faces"
296 sAvgSubjFacesSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'WAvg: Avg: Faces');
297 % Process: Select file comments with tag: "WAvg: Avg: Scrambled"
298 sAvgSubjScrambledSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'WAvg: Avg: Scrambled');
299 % Process: Difference: A-B
300 sDiffFacesSubjSrcMeg = bst_process('CallProcess', 'process_diff_ab', sAvgSubjFacesSrcMeg, sAvgSubjScrambledSrcMeg, ...
301 'source_abs', 0);
302 % Process: Set comment
303 sDiffFacesSubjSrcMeg = bst_process('CallProcess', 'process_set_comment', sDiffFacesSubjSrcMeg, [], ...
304 'tag', 'Faces - Scrambled | MEG', ...
305 'isindex', 0);
306
307 % ===== FAMOUS - UNFAMILIAR: SOURCE EEG =====
308 % Process: Select file comments with tag: "WAvg: Avg: Famous"
309 sAvgSubjFamousSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Famous');
310 % Process: Select file comments with tag: "WAvg: Avg: Unfamiliar"
311 sAvgSubjUnfamiliarSrcEeg = bst_process('CallProcess', 'process_select_tag', sSrcEeg, [], 'tag', 'WAvg: Avg: Unfamiliar');
312 % Process: Difference: A-B
313 sDiffFamousSubjSrcEeg = bst_process('CallProcess', 'process_diff_ab', sAvgSubjFamousSrcEeg, sAvgSubjUnfamiliarSrcEeg, ...
314 'source_abs', 0);
315 % Process: Set comment
316 sDiffFamousSubjSrcEeg = bst_process('CallProcess', 'process_set_comment', sDiffFamousSubjSrcEeg, [], ...
317 'tag', 'Famous - Unfamiliar | EEG', ...
318 'isindex', 0);
319
320 % ===== FAMOUS - UNFAMILIAR: SOURCE MEG =====
321 % Process: Select file comments with tag: "WAvg: Avg: Famous"
322 sAvgSubjFamousSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'WAvg: Avg: Famous');
323 % Process: Select file comments with tag: "WAvg: Avg: Unfamiliar"
324 sAvgSubjUnfamiliarSrcMeg = bst_process('CallProcess', 'process_select_tag', sSrcMeg, [], 'tag', 'WAvg: Avg: Unfamiliar');
325 % Process: Difference: A-B
326 sDiffFamousSubjSrcMeg = bst_process('CallProcess', 'process_diff_ab', sAvgSubjFamousSrcMeg, sAvgSubjUnfamiliarSrcMeg, ...
327 'source_abs', 0);
328 % Process: Set comment
329 sDiffFamousSubjSrcMeg = bst_process('CallProcess', 'process_set_comment', sDiffFamousSubjSrcMeg, [], ...
330 'tag', 'Famous - Unfamiliar | MEG', ...
331 'isindex', 0);
332
333
334
335 %% ===== SUBJECT AVERAGES: FILTER AND NORMALIZE =======================================
336 % ====================================================================================
337
338 % ===== FILTER: MEG+EEG =====
339 % Process: Select data files in: */Intra
340 sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
341 'subjectname', 'All', ...
342 'condition', DirIntra, ...
343 'includeintra', 1);
344 % Process: Low-pass:32Hz
345 sAvgData = bst_process('CallProcess', 'process_bandpass', sAvgData, [], ...
346 'sensortypes', 'MEG, EEG', ...
347 'highpass', 0, ...
348 'lowpass', 32, ...
349 'attenuation', 'strict', ... % 60dB
350 'mirror', 0, ...
351 'overwrite', 1);
352 % Process: Extract time: [-202ms,900ms]
353 sAvgData = bst_process('CallProcess', 'process_extract_time', sAvgData, [], ...
354 'timewindow', [-0.2018, 0.9], ...
355 'overwrite', 1);
356
357 % ===== FILTER AND NORMALIZE: SOURCES =====
358 % Process: Select source files in: */Intra
359 sAvgSrc = bst_process('CallProcess', 'process_select_files_results', [], [], ...
360 'subjectname', 'All', ...
361 'condition', DirIntra, ...
362 'includeintra', 1);
363 % Process: Low-pass:32Hz
364 sAvgSrc = bst_process('CallProcess', 'process_bandpass', sAvgSrc, [], ...
365 'highpass', 0, ...
366 'lowpass', 32, ...
367 'attenuation', 'strict', ... % 60dB
368 'mirror', 0, ...
369 'overwrite', 1);
370 % Process: Extract time: [-202ms,900ms]
371 sAvgSrc = bst_process('CallProcess', 'process_extract_time', sAvgSrc, [], ...
372 'timewindow', [-0.2018, 0.9], ...
373 'overwrite', 1);
374 % Process: Z-score transformation: [-202ms,-5ms]
375 sAvgSrc = bst_process('CallProcess', 'process_baseline_norm', sAvgSrc, [], ...
376 'baseline', [-0.2018, -0.005], ...
377 'source_abs', 0, ...
378 'method', 'zscore', ... % Z-score transformation: x_std = (x - μ) / σ
379 'overwrite', 1);
380
381 % ===== NORMALIZE: TIMEFREQ =====
382 % Process: Select time-frequency files in: */Intra
383 sAvgTf = bst_process('CallProcess', 'process_select_files_timefreq', [], [], ...
384 'subjectname', 'All', ...
385 'condition', DirIntra, ...
386 'includeintra', 1);
387 % Process: Event-related perturbation (ERS/ERD): [-200ms,-4ms]
388 sAvgTf = bst_process('CallProcess', 'process_baseline_norm', sAvgTf, [], ...
389 'baseline', [-0.2, -0.004], ...
390 'method', 'ersd', ... % Event-related perturbation (ERS/ERD): x_std = (x - μ) / μ * 100
391 'overwrite', 1);
392
393 % Save report
394 ReportFile = bst_report('Save', []);
395 if ~isempty(reports_dir) && ~isempty(ReportFile)
396 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolName '_1prepare.html']));
397 end
398
399
400
401 %% ===== SUBJECT AVERAGES: SCREEN CAPTURES ============================================
402 % ====================================================================================
403 % Start a new report
404 bst_report('Start');
405 % Set colormap: local color scale
406 bst_colormaps('SetMaxMode', 'stat2', 'local');
407
408 % ===== FACES: RECORDINGS ======
409 % Process: Select data files in: */*/WAvg: Avg: Faces |
410 sAvgFaces = bst_process('CallProcess', 'process_select_files_data', [], [], ...
411 'subjectname', 'All', ...
412 'tag', 'WAvg: Avg: Faces |', ...
413 'includeintra', 1);
414 % Process: Snapshot: Recordings time series
415 bst_process('CallProcess', 'process_snapshot', sAvgFaces, [], ...
416 'target', 1, ... % Sensor/MRI registration
417 'modality', 1); % MEG
418 % Process: Snapshot: Recordings time series
419 bst_process('CallProcess', 'process_snapshot', sAvgFaces, [], ...
420 'target', 5, ... % Recordings time series
421 'modality', 4, ... % EEG
422 'time', 0.1055);
423
424 % ===== FACES: SOURCES MEG ======
425 % Process: Select source files in: */*/WAvg: Avg: Faces | MEG
426 sAvgFacesSrcMeg = bst_process('CallProcess', 'process_select_files_results', [], [], ...
427 'subjectname', 'All', ...
428 'condition', DirIntra, ...
429 'tag', 'WAvg: Avg: Faces | MEG', ...
430 'includeintra', 1);
431 % Process: Snapshot: Sources (one time)
432 bst_process('CallProcess', 'process_snapshot', sAvgFacesSrcMeg, [], ...
433 'target', 8, ... % Sources (one time)
434 'orient', 6, ... % back
435 'time', 0.1055, ...
436 'threshold', 10);
437
438 % ===== FACES: TIME-FREQ EEG ======
439 % Process: Select time-frequency files in: */*/WAvg: Avg: Faces
440 sAvgFacesTf = bst_process('CallProcess', 'process_select_files_timefreq', [], [], ...
441 'subjectname', 'All', ...
442 'tag', 'WAvg: Avg: Faces', ...
443 'includeintra', 1);
444 % Process: Snapshot: Time-frequency maps
445 bst_process('CallProcess', 'process_snapshot', sAvgFacesTf, [], ...
446 'target', 14, ... % Time-frequency maps
447 'time', 0.1055, ...
448 'rowname', 'EEG070');
449
450 % Save report
451 ReportFile = bst_report('Save', []);
452 if ~isempty(reports_dir) && ~isempty(ReportFile)
453 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolName '_2snapshots.html']));
454 end
455
456
457
458 %% ===== GROUP ANALYSIS: FACES-SCRAMBLED: MEG/EEG =====================================
459 % ====================================================================================
460 % Start a new report
461 bst_report('Start');
462 % Set colormap: global color scale
463 bst_colormaps('SetMaxMode', 'meg', 'global');
464 bst_colormaps('SetMaxMode', 'eeg', 'global');
465 % Set display properties for statistics: p<0.05, FDR-corrected
466 StatThreshOptions = bst_get('StatThreshOptions');
467 StatThreshOptions.pThreshold = 0.05;
468 StatThreshOptions.Correction = 'fdr';
469 StatThreshOptions.Control = [1 2 3];
470 bst_set('StatThreshOptions', StatThreshOptions);
471
472 % ===== GRAND AVERAGES =====
473 % Process: Select data files in: Group_analysis/Intra
474 sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
475 'subjectname', GroupSubjectName, ...
476 'condition', DirIntra, ...
477 'includeintra', 1);
478 % Take screen captures
479 DataScreenCapture(sAvgData(1), 1, 1, '');
480 DataScreenCapture(sAvgData(2), 1, 1, '');
481 DataScreenCapture(sAvgData(3), 1, 1, '');
482 DataScreenCapture(sAvgData(4), 1, 1, '');
483 % Define function for screen captures
484 function DataScreenCapture(sFiles, isMEG, isEEG, Comment)
485 if isMEG
486 % Process: Snapshot: Recordings time series
487 bst_process('CallProcess', 'process_snapshot', sFiles, [], ...
488 'target', 5, ... % Recordings time series
489 'modality', 3, ... % MEG (Magnetometers)
490 'time', 0.1055, ...
491 'Comment', ['MEG MAG: ' Comment]);
492 % Process: Snapshot: Recordings topography (contact sheet)
493 bst_process('CallProcess', 'process_snapshot', sFiles, [], ...
494 'target', 7, ... % Recordings topography (contact sheet)
495 'modality', 3, ... % MEG (Magnetometers)
496 'contact_time', [0.050, 0.300], ...
497 'contact_nimage', 6, ...
498 'Comment', ['MEG MAG: ' Comment]);
499 end
500 if isEEG
501 % Process: Snapshot: Recordings time series
502 bst_process('CallProcess', 'process_snapshot', sFiles, [], ...
503 'target', 5, ... % Recordings time series
504 'modality', 4, ... % EEG
505 'time', 0.1055, ...
506 'Comment', ['EEG: ' Comment]);
507 % Process: Snapshot: Recordings topography (contact sheet)
508 bst_process('CallProcess', 'process_snapshot', sFiles, [], ...
509 'target', 7, ... % Recordings topography (contact sheet)
510 'modality', 4, ... % EEG
511 'contact_time', [0.050, 0.300], ...
512 'contact_nimage', 6, ...
513 'Comment', ['EEG: ' Comment]);
514 end
515 end
516
517 % ===== FACES-SCRAMBLED: MEAN DIFF =====
518 % Process: Select data files in: */DirIntra/"Avg: Faces | "
519 sSubjAvgFacesData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
520 'subjectname', 'All', ...
521 'condition', DirIntra, ...
522 'tag', 'Avg: Faces | ', ...
523 'includeintra', 1);
524 % Process: Select data files in: */DirIntra/"WAvg: Avg: Scrambled (6 files)"
525 sSubjAvgScrambledData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
526 'subjectname', 'All', ...
527 'condition', DirIntra, ...
528 'tag', 'WAvg: Avg: Scrambled (6 files)', ...
529 'includeintra', 1);
530 % Process: Difference of means [mean]
531 sDiffFacesData = bst_process('CallProcess', 'process_diff_mean', sSubjAvgFacesData, sSubjAvgScrambledData, ...
532 'avg_func', 1, ... % Arithmetic average mean(A) - mean(B)
533 'weighted', 0);
534 % Process: Set comment
535 sDiffFacesData = bst_process('CallProcess', 'process_set_comment', sDiffFacesData, [], ...
536 'tag', 'Faces - Scrambled', ...
537 'isindex', 0);
538 % Take screen captures
539 DataScreenCapture(sDiffFacesData, 1, 1, sDiffFacesData.Comment);
540
541 % ===== FACES-SCRAMBLED: PARAMETRIC T-TEST =====
542 % Process: Select data files in: */DirIntra/"Avg: Faces | "
543 sSubjAvgFacesData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
544 'subjectname', 'All', ...
545 'condition', DirIntra, ...
546 'tag', 'Avg: Faces | ', ...
547 'includeintra', 1);
548 % Process: Select data files in: */DirIntra/"WAvg: Avg: Scrambled (6 files)"
549 sSubjAvgScrambledData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
550 'subjectname', 'All', ...
551 'condition', DirIntra, ...
552 'tag', 'WAvg: Avg: Scrambled (6 files)', ...
553 'includeintra', 1);
554 % Process: t-test paired [ALL] H0:(A=B), H1:(A<>B)
555 sStatParamFacesData = bst_process('CallProcess', 'process_test_parametric2p', sSubjAvgFacesData, sSubjAvgScrambledData, ...
556 'timewindow', [], ...
557 'sensortypes', '', ...
558 'isabs', 0, ...
559 'avgtime', 0, ...
560 'avgrow', 0, ...
561 'Comment', '', ...
562 'test_type', 'ttest_paired', ... % Paired Student's t-test (A-B)~N(m,v)t = mean(A-B) / std(A-B) * sqrt(n) df=n-1
563 'tail', 'two'); % Two-tailed
564 % Process: Set comment
565 sStatParamFacesData = bst_process('CallProcess', 'process_set_comment', sStatParamFacesData, [], ...
566 'tag', 'Faces - Scrambled: Parametric t-test', ...
567 'isindex', 0);
568 % Take screen captures
569 DataScreenCapture(sStatParamFacesData, 1, 1, sStatParamFacesData.Comment);
570
571 % ===== FACES-SCRAMBLED: NON-PARAMETRIC T-TEST =====
572 % Process: Perm t-test paired [All] H0:(A=B), H1:(A<>B)
573 sStatPermFacesData = bst_process('CallProcess', 'process_test_permutation2p', sSubjAvgFacesData, sSubjAvgScrambledData, ...
574 'timewindow', [], ...
575 'sensortypes', '', ...
576 'isabs', 0, ...
577 'avgtime', 0, ...
578 'avgrow', 0, ...
579 'iszerobad', 0, ...
580 'Comment', '', ...
581 'test_type', 'ttest_paired', ... % Paired Student's t-test T = mean(A-B) / std(A-B) * sqrt(n)
582 'randomizations', 1000, ...
583 'tail', 'two'); % Two-tailed
584 % Process: Set comment
585 sStatPermFacesData = bst_process('CallProcess', 'process_set_comment', sStatPermFacesData, [], ...
586 'tag', 'Faces - Scrambled: Permutation t-test', ...
587 'isindex', 0);
588 % Take screen captures
589 DataScreenCapture(sStatPermFacesData, 1, 1, sStatPermFacesData.Comment);
590
591 % ===== FACES-SCRAMBLED: CLUSTER T-TEST =====
592 % Process: FT t-test paired cluster [all EEG] H0:(A=B), H1:(A<>B)
593 sStatClustFacesData = bst_process('CallProcess', 'process_ft_timelockstatistics', sSubjAvgFacesData, sSubjAvgScrambledData, ...
594 'sensortypes', 'EEG', ...
595 'timewindow', [], ...
596 'isabs', 0, ...
597 'avgtime', 0, ...
598 'avgchan', 0, ...
599 'randomizations', 1000, ...
600 'statistictype', 2, ... % Paired t-test
601 'tail', 'two', ... % Two-tailed
602 'correctiontype', 2, ... % cluster
603 'minnbchan', 0, ...
604 'clusteralpha', 0.05);
605 % If FieldTrip is available and results were returned
606 if ~isempty(sStatClustFacesData)
607 % Process: Set comment
608 sStatClustFacesData = bst_process('CallProcess', 'process_set_comment', sStatClustFacesData, [], ...
609 'tag', 'Faces - Scrambled: Cluster t-test EEG', ...
610 'isindex', 0);
611 % Take screen captures
612 DataScreenCapture(sStatClustFacesData, 0, 1, sStatClustFacesData.Comment);
613 end
614
615
616 %% ===== GROUP ANALYSIS: FACES-SCRAMBLED: MEG/EEG =====================================
617 % ====================================================================================
618 % ===== FAMOUS-UNFAMILIAR: MEAN DIFF =====
619 % Process: Select data files in: */DirIntra/"Avg: Famous | "
620 sSubjAvgFamousData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
621 'subjectname', 'All', ...
622 'condition', DirIntra, ...
623 'tag', 'Avg: Famous (6 files)', ...
624 'includeintra', 1);
625 % Process: Select data files in: */DirIntra/"WAvg: Avg: Unfamiliar (6 files)"
626 sSubjAvgUnfamiliarData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
627 'subjectname', 'All', ...
628 'condition', DirIntra, ...
629 'tag', 'WAvg: Avg: Unfamiliar (6 files)', ...
630 'includeintra', 1);
631 % Process: Difference of means [mean]
632 sDiffFamousData = bst_process('CallProcess', 'process_diff_mean', sSubjAvgFamousData, sSubjAvgUnfamiliarData, ...
633 'avg_func', 1, ... % Arithmetic average mean(A) - mean(B)
634 'weighted', 0);
635 % Process: Set comment
636 sDiffFamousData = bst_process('CallProcess', 'process_set_comment', sDiffFamousData, [], ...
637 'tag', 'Famous - Unfamiliar', ...
638 'isindex', 0);
639 % Take screen captures
640 DataScreenCapture(sDiffFamousData, 1, 1, sDiffFamousData.Comment);
641
642 % ===== FAMOUS-UNFAMILIAR: PARAMETRIC T-TEST =====
643 % Process: t-test paired [All] H0:(A=B), H1:(A<>B)
644 sStatParamFamousData = bst_process('CallProcess', 'process_test_parametric2p', sSubjAvgFamousData, sSubjAvgUnfamiliarData, ...
645 'timewindow', [], ...
646 'sensortypes', '', ...
647 'isabs', 0, ...
648 'avgtime', 0, ...
649 'avgrow', 0, ...
650 'Comment', '', ...
651 'test_type', 'ttest_paired', ... % Paired Student's t-test (A-B)~N(m,v)t = mean(A-B) / std(A-B) * sqrt(n) df=n-1
652 'tail', 'two'); % Two-tailed
653 % Process: Set comment
654 sStatParamFamousData = bst_process('CallProcess', 'process_set_comment', sStatParamFamousData, [], ...
655 'tag', 'Famous - Unfamiliar: Parametric t-test', ...
656 'isindex', 0);
657 % Take screen captures
658 DataScreenCapture(sStatParamFamousData, 1, 1, sStatParamFamousData.Comment);
659
660 % ===== FAMOUS-UNFAMILIAR: CLUSTER T-TEST =====
661 % Process: FT t-test paired cluster [all EEG] H0:(A=B), H1:(A<>B)
662 sStatClustFamousData = bst_process('CallProcess', 'process_ft_timelockstatistics', sSubjAvgFamousData, sSubjAvgUnfamiliarData, ...
663 'sensortypes', 'EEG', ...
664 'timewindow', [], ...
665 'isabs', 0, ...
666 'avgtime', 0, ...
667 'avgchan', 0, ...
668 'randomizations', 1000, ...
669 'statistictype', 2, ... % Paired t-test
670 'tail', 'two', ... % Two-tailed
671 'correctiontype', 2, ... % cluster
672 'minnbchan', 0, ...
673 'clusteralpha', 0.05);
674 % If FieldTrip is available and results were returned
675 if ~isempty(sStatClustFacesData)
676 % Process: Set comment
677 sStatClustFamousData = bst_process('CallProcess', 'process_set_comment', sStatClustFamousData, [], ...
678 'tag', 'Famous - Unfamiliar: Cluster t-test EEG', ...
679 'isindex', 0);
680 % Take screen captures
681 DataScreenCapture(sStatClustFamousData, 0, 1, sStatClustFamousData.Comment);
682 end
683
684 % Save report
685 ReportFile = bst_report('Save', []);
686 if ~isempty(reports_dir) && ~isempty(ReportFile)
687 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolName '_3meeg.html']));
688 end
689
690
691
692 %% ===== FACES-SCRAMBLED: SOURCES (MEG) ===============================================
693 % ====================================================================================
694 % Start a new report
695 bst_report('Start');
696
697 % ===== PROJECT SOURCES =====
698 % Process: Select source files in: */Intra
699 sAvgSrc = bst_process('CallProcess', 'process_select_files_results', [], [], ...
700 'subjectname', 'All', ...
701 'condition', DirIntra, ...
702 'includeintra', 1);
703 % Process: Absolute values / Norm for unconstrained
704 if isUnconstrained
705 % Process: Unconstrained to flat map
706 sAvgSrc = bst_process('CallProcess', 'process_source_flat', sAvgSrc, [], ...
707 'method', 1); % Norm: sqrt(x^2+y^2+z^2)
708 else
709 sAvgSrc = bst_process('CallProcess', 'process_absolute', sAvgSrc, [], ...
710 'overwrite', 1);
711 end
712 % Process: Project on default anatomy
713 sProjSrc = bst_process('CallProcess', 'process_project_sources', sAvgSrc, [], ...
714 'headmodeltype', 'surface'); % Cortex surface
715 % Process: Spatial smoothing (3.00,abs)
716 sProjSrc = bst_process('CallProcess', 'process_ssmooth_surfstat', sProjSrc, [], ...
717 'fwhm', 3, ...
718 'overwrite', 1, ...
719 'source_abs', 1);
720
721 % ===== MOVE FILES =====
722 % File tags / destination folder
723 ListTags = {'Avg: Faces | EEG', 'Faces_EEG'; ...
724 'Avg: Faces | MEG', 'Faces_MEG'; ...
725 'Avg: Famous (6 files) | EEG', 'Famous_EEG'; ...
726 'Avg: Famous (6 files) | MEG', 'Famous_MEG'; ...
727 'Avg: Unfamiliar (6 files) | EEG', 'Unfamiliar_EEG'; ...
728 'Avg: Unfamiliar (6 files) | MEG', 'Unfamiliar_MEG'; ...
729 'Avg: Scrambled (6 files) | EEG', 'Scrambled_EEG'; ...
730 'Avg: Scrambled (6 files) | MEG', 'Scrambled_MEG'; ...
731 'Faces - Scrambled | EEG', 'Faces-Scrambled_EEG'; ...
732 'Faces - Scrambled | MEG', 'Faces-Scrambled_MEG'; ...
733 'Famous - Unfamiliar | EEG', 'Famous-Unfamiliar_EEG'; ...
734 'Famous - Unfamiliar | MEG', 'Famous-Unfamiliar_MEG'};
735 % Move group by group
736 for i = 1:size(ListTags,1)
737 % Find names of files to move
738 iTag = find(~cellfun(@(c)isempty(strfind(c, ListTags{i,1})), {sProjSrc.Comment}));
739 if isempty(iTag)
740 continue;
741 end
742 % Process: Move files
743 bst_process('CallProcess', 'process_movefile', {sProjSrc(iTag).FileName}, [], ...
744 'subjectname', GroupSubjectName, ...
745 'folder', ListTags{i,2});
746 end
747
748
749 % ===== LOOP ON MODALITY =====
750 AllModalities = {'MEG','EEG'};
751 for iMod = 1:length(AllModalities)
752 % Selected modality for this loop
753 Mod = AllModalities{iMod};
754
755 % ===== |FACES-SCRAMBLED|: MEAN DIFF ======
756 % Process: Select source files in: Group_analysis/Faces-Scrambled_MOD
757 sDiffFaces = bst_process('CallProcess', 'process_select_files_results', [], [], ...
758 'subjectname', GroupSubjectName, ...
759 'condition', ['Faces-Scrambled_', Mod]);
760 % Process: Weighted Average: By folder (grand average)
761 sAbsDiffFaces = bst_process('CallProcess', 'process_average', sDiffFaces, [], ...
762 'avgtype', 4, ... % By folder (grand average)
763 'avg_func', 1, ... % Arithmetic average: mean(x)
764 'weighted', 0, ...
765 'scalenormalized', 0);
766 % Process: Set comment
767 sAbsDiffFaces = bst_process('CallProcess', 'process_set_comment', sAbsDiffFaces, [], ...
768 'tag', ['mean(|Faces-Scrambled|) | ' Mod], ...
769 'isindex', 0);
770 % Process: Move files
771 sAbsDiffFaces = bst_process('CallProcess', 'process_movefile', sAbsDiffFaces, [], ...
772 'subjectname', GroupSubjectName, ...
773 'folder', DirIntra);
774 % Colormap: Custom max: [-10,+10] Z
775 bst_colormaps('SetMaxCustom', 'stat2', [], -10, 10);
776 % Process: Snapshot: Sources (contact sheet)
777 bst_process('CallProcess', 'process_snapshot', sAbsDiffFaces, [], ...
778 'target', 9, ... % Sources (contact sheet)
779 'orient', 4, ... % bottom
780 'contact_time', [0.05, 0.4], ...
781 'contact_nimage', 16, ...
782 'threshold', 30, ...
783 'Comment', [Mod ': mean(|Faces-Scrambled|)']);
784 % Process: Snapshot: Sources (contact sheet)
785 bst_process('CallProcess', 'process_snapshot', sAbsDiffFaces, [], ...
786 'target', 9, ... % Sources (contact sheet)
787 'orient', 2, ... % right
788 'contact_time', [0.05, 0.4], ...
789 'contact_nimage', 16, ...
790 'threshold', 30, ...
791 'Comment', [Mod ': mean(|Faces-Scrambled|)']);
792
793 % ===== |FACES-SCRAMBLED|: PARAMETRIC CHI2-TEST ======
794 % Process: Select source files in: Group_analysis/Faces-Scrambled_MOD
795 sDiffFaces = bst_process('CallProcess', 'process_select_files_results', [], [], ...
796 'subjectname', GroupSubjectName, ...
797 'condition', ['Faces-Scrambled_' Mod]);
798 % Process: Chi2-test [all] H0:(|Zi| = 0)
799 sChiParamFaces = bst_process('CallProcess', 'process_test_parametric1', sDiffFaces, [], ...
800 'timewindow', [], ...
801 'scoutsel', {}, ...
802 'scoutfunc', 1, ... % Mean
803 'isnorm', 0, ...
804 'avgtime', 0, ...
805 'Comment', '', ...
806 'test_type', 'chi2_onesample', ... % One-sample Chi2 test Zi~N(0,1), i=1..nQ = sum(|Zi|^2) Q~Chi2(n)
807 'tail', 'two'); % Two-tailed
808 % Process: Set comment
809 sChiParamFaces = bst_process('CallProcess', 'process_set_comment', sChiParamFaces, [], ...
810 'tag', ['|Faces-Scrambled|=0: Parametric Chi2 test | ' Mod], ...
811 'isindex', 0);
812 % Process: Move files
813 sChiParamFaces = bst_process('CallProcess', 'process_movefile', sChiParamFaces, [], ...
814 'subjectname', GroupSubjectName, ...
815 'folder', DirIntra);
816 % Set colormap: global color scale
817 bst_colormaps('SetMaxMode', 'stat2', 'global');
818 % Process: Snapshot: Sources (contact sheet)
819 bst_process('CallProcess', 'process_snapshot', sChiParamFaces, [], ...
820 'target', 9, ... % Sources (contact sheet)
821 'orient', 4, ... % bottom
822 'contact_time', [0.05, 0.4], ...
823 'contact_nimage', 16, ...
824 'Comment', [Mod ': |Faces-Scrambled|=0: Parametric Chi2-test']);
825 % Process: Snapshot: Sources (contact sheet)
826 bst_process('CallProcess', 'process_snapshot', sChiParamFaces, [], ...
827 'target', 9, ... % Sources (contact sheet)
828 'orient', 2, ... % right
829 'contact_time', [0.05, 0.4], ...
830 'contact_nimage', 16, ...
831 'Comment', [Mod ': |Faces-Scrambled|=0: Parametric Chi2-test']);
832
833 % ===== LOG(|FACES-SCRAMBLED|): PARAMETRIC CHI2-TEST ======
834 % Process: Select source files in: Group_analysis/Faces-Scrambled_MOD
835 sDiffFaces = bst_process('CallProcess', 'process_select_files_results', [], [], ...
836 'subjectname', GroupSubjectName, ...
837 'condition', ['Faces-Scrambled_' Mod]);
838 % Process: Duplicate folders: Add tag "_log"
839 sDiffFacesLog = bst_process('CallProcess', 'process_duplicate', sDiffFaces, [], ...
840 'target', 2, ... % Duplicate folders
841 'tag', '_log');
842 % Process: Run Matlab command
843 sDiffFacesLog = bst_process('CallProcess', 'process_matlab_eval', sDiffFacesLog, [], ...
844 'matlab', 'Data = log(Data);', ...
845 'overwrite', 1);
846 % Process: Chi2-test [all] H0:(|Zi| = 0)
847 sChiParamFacesLog = bst_process('CallProcess', 'process_test_parametric1', sDiffFacesLog, [], ...
848 'timewindow', [], ...
849 'scoutsel', {}, ...
850 'scoutfunc', 1, ... % Mean
851 'isnorm', 0, ...
852 'avgtime', 0, ...
853 'Comment', '', ...
854 'test_type', 'chi2_onesample', ... % One-sample Chi2 test Zi~N(0,1), i=1..nQ = sum(|Zi|^2) Q~Chi2(n)
855 'tail', 'two'); % Two-tailed
856 % Process: Set comment
857 sChiParamFacesLog = bst_process('CallProcess', 'process_set_comment', sChiParamFacesLog, [], ...
858 'tag', ['log(|Faces-Scrambled|)=0: Parametric Chi2 test | ' Mod], ...
859 'isindex', 0);
860 % Process: Move files
861 sChiParamFacesLog = bst_process('CallProcess', 'process_movefile', sChiParamFacesLog, [], ...
862 'subjectname', GroupSubjectName, ...
863 'folder', DirIntra);
864 % Set colormap: global color scale
865 bst_colormaps('SetMaxMode', 'stat2', 'global');
866 % Process: Snapshot: Sources (contact sheet)
867 bst_process('CallProcess', 'process_snapshot', sChiParamFacesLog, [], ...
868 'target', 9, ... % Sources (contact sheet)
869 'orient', 4, ... % bottom
870 'contact_time', [0.05, 0.4], ...
871 'contact_nimage', 16, ...
872 'Comment', [Mod ': log(|Faces-Scrambled|)=0: Parametric Chi2-test']);
873 % Process: Snapshot: Sources (contact sheet)
874 bst_process('CallProcess', 'process_snapshot', sChiParamFacesLog, [], ...
875 'target', 9, ... % Sources (contact sheet)
876 'orient', 2, ... % right
877 'contact_time', [0.05, 0.4], ...
878 'contact_nimage', 16, ...
879 'Comment', [Mod ': log(|Faces-Scrambled|)=0: Parametric Chi2-test']);
880
881
882 % ===== |FACES|-|SCRAMBLED|: MEAN DIFF ======
883 % Process: Select source files in: Group_analysis/Faces_MOD
884 sAvgFaces = bst_process('CallProcess', 'process_select_files_results', [], [], ...
885 'subjectname', GroupSubjectName, ...
886 'condition', ['Faces_' Mod]);
887 % Process: Select source files in: Group_analysis/Scrambled_MOD
888 sAvgScrambled = bst_process('CallProcess', 'process_select_files_results', [], [], ...
889 'subjectname', GroupSubjectName, ...
890 'condition', ['Scrambled_' Mod]);
891 % Process: Difference of means [abs(mean)]
892 sDiffAbsFaces = bst_process('CallProcess', 'process_diff_mean', sAvgFaces, sAvgScrambled, ...
893 'avg_func', 2, ... % Absolute value of average abs(mean(A)) - abs(mean(B))
894 'weighted', 0);
895 % Process: Set comment
896 sDiffAbsFaces = bst_process('CallProcess', 'process_set_comment', sDiffAbsFaces, [], ...
897 'tag', ['mean(|Faces|)-mean(|Scrambled|) | ' Mod], ...
898 'isindex', 0);
899 % Colormap: Custom max: [-10,+10] Z
900 bst_colormaps('SetMaxCustom', 'stat2', [], -10, 10);
901 % Process: Snapshot: Sources (contact sheet)
902 bst_process('CallProcess', 'process_snapshot', sDiffAbsFaces, [], ...
903 'target', 9, ... % Sources (contact sheet)
904 'orient', 4, ... % bottom
905 'contact_time', [0.05, 0.4], ...
906 'contact_nimage', 16, ...
907 'threshold', 30, ...
908 'Comment', [Mod ': mean(|Faces|)-mean(|Scrambled|)']);
909 % Process: Snapshot: Sources (contact sheet)
910 bst_process('CallProcess', 'process_snapshot', sDiffAbsFaces, [], ...
911 'target', 9, ... % Sources (contact sheet)
912 'orient', 2, ... % right
913 'contact_time', [0.05, 0.4], ...
914 'contact_nimage', 16, ...
915 'threshold', 30, ...
916 'Comment', [Mod ': mean(|Faces|)-mean(|Scrambled|)']);
917
918 % ===== |FACES|-|SCRAMBLED|: PARAMETRIC T-TEST ======
919 % Process: Select source files in: Group_analysis/Faces_MOD
920 sAvgFaces = bst_process('CallProcess', 'process_select_files_results', [], [], ...
921 'subjectname', GroupSubjectName, ...
922 'condition', ['Faces_' Mod]);
923 % Process: Select source files in: Group_analysis/Scrambled_MOD
924 sAvgScrambled = bst_process('CallProcess', 'process_select_files_results', [], [], ...
925 'subjectname', GroupSubjectName, ...
926 'condition', ['Scrambled_' Mod]);
927 % Process: t-test paired [-202ms,900ms] H0:(A=B), H1:(A<>B)
928 sTtestParamFaces = bst_process('CallProcess', 'process_test_parametric2p', sAvgFaces, sAvgScrambled, ...
929 'timewindow', [], ...
930 'scoutsel', {}, ...
931 'scoutfunc', 1, ... % Mean
932 'isnorm', 0, ...
933 'avgtime', 0, ...
934 'Comment', '', ...
935 'test_type', 'ttest_paired', ... % Paired Student's t-test (A-B)~N(m,v)t = mean(A-B) / std(A-B) * sqrt(n) df=n-1
936 'tail', 'two'); % Two-tailed
937 % Process: Set comment
938 sTtestParamFaces = bst_process('CallProcess', 'process_set_comment', sTtestParamFaces, [], ...
939 'tag', ['|Faces|=|Scrambled|: Parametric t-test | ' Mod], ...
940 'isindex', 0);
941 % Set colormap: global color scale
942 bst_colormaps('SetMaxMode', 'stat2', 'global');
943 % Process: Snapshot: Sources (contact sheet)
944 bst_process('CallProcess', 'process_snapshot', sTtestParamFaces, [], ...
945 'target', 9, ... % Sources (contact sheet)
946 'orient', 4, ... % bottom
947 'contact_time', [0.05, 0.4], ...
948 'contact_nimage', 16, ...
949 'Comment', [Mod ': |Faces|-|Scrambled|: Parametric t-test']);
950 % Process: Snapshot: Sources (contact sheet)
951 bst_process('CallProcess', 'process_snapshot', sTtestParamFaces, [], ...
952 'target', 9, ... % Sources (contact sheet)
953 'orient', 2, ... % right
954 'contact_time', [0.05, 0.4], ...
955 'contact_nimage', 16, ...
956 'Comment', [Mod ': |Faces|-|Scrambled|: Parametric t-test']);
957 end
958
959
960
961 % Save report
962 ReportFile = bst_report('Save', []);
963 if ~isempty(reports_dir) && ~isempty(ReportFile)
964 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolName '_4sources.html']));
965 end
966
967
968
969
970 end
971