Hi all, I have a question about the order of the vertices in the surface file. Is it related to the Brodmann area? if yes, can you give me a table or a list of the order relative to the region( Brodmann area)? if no, the order of the vertices is based on what?
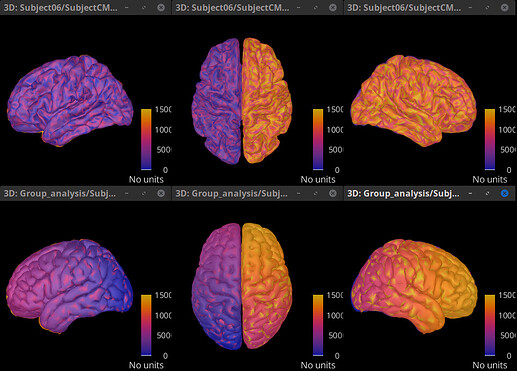
The order of vertices will be assigned by the MRI segmentation software. It does not seems to be related to the anatomy. See the figures below, where the colorbar indicates the vertex number, (top) vertex order for the FSAverage, (bottom) vertex order for ICBM152 processed with FreeSurfer. Both with 15002 vertices.
Is there a specific reason why the vertex order is relevant to you?
The order of the vertices does matter, however, for computations purposes,
it's optimized to have the nodes ordered to be neighbors.
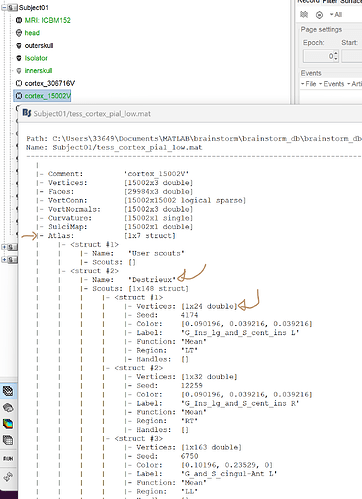
The scout's vertices are saved within the surface file:
Right-click on the cortex => file => view file contents (see attached picture)
For each scout, you have the list of the associated vertices as well as all the information related to the scouts.
I am trying to analyze some regions with different w files, so if the order of the vertices is related to specific regions, it will save my time by comparing those vertices directly.
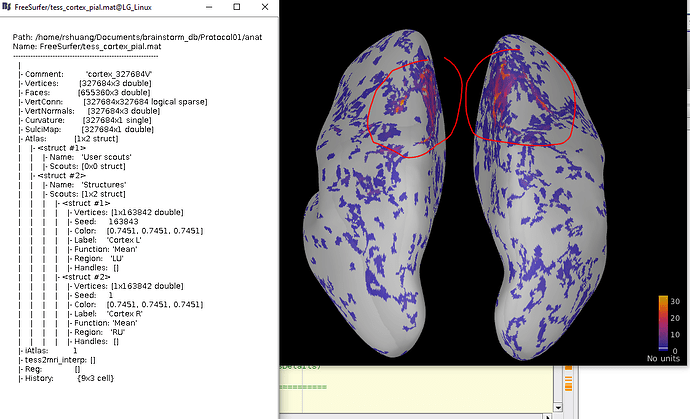
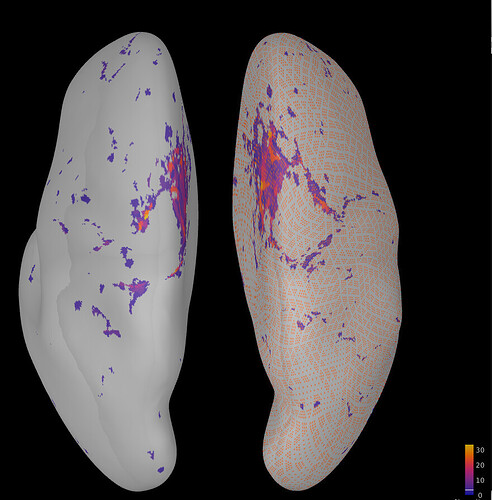
Thanks your reply. But in my case, my cortex is only merged by left brain surface file and right brain surface file, so the scount is [1*2 struct]. What I am trying to do is find the regions that related to the order of the 163942 vertices. For example, the red-circled regions may be a set of serial numbers in vertices in order (eg: 1000-2000).
I modified the w file [set 1:20000 vertices be 25] to see the difference, and the result is that
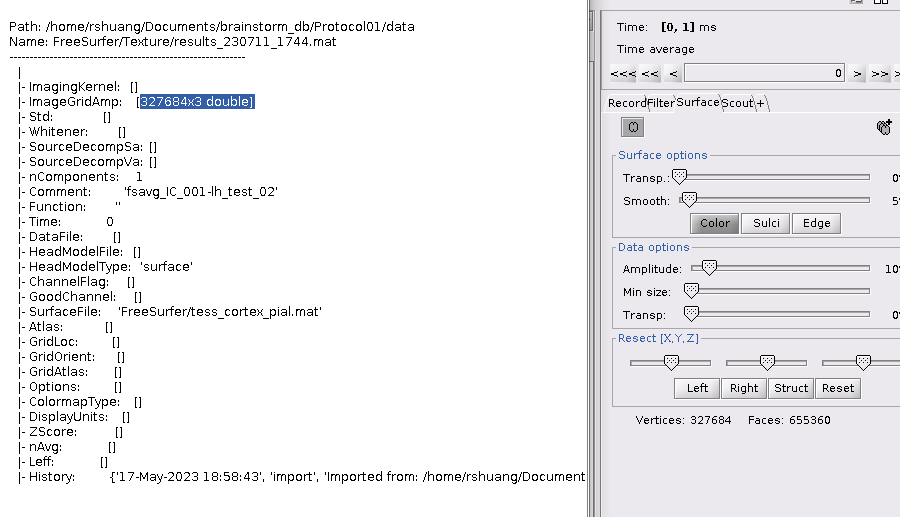
, It shows that I cannot compare w files directly based on the vertices' order. On the other hand, you told me that the struct of ImageGridAmp can also be used in time, eg: [327684x2 double] have [0,1] ms, but when I added one more column, to form [327684x3 double], the time option still only have [0,1], why is that?
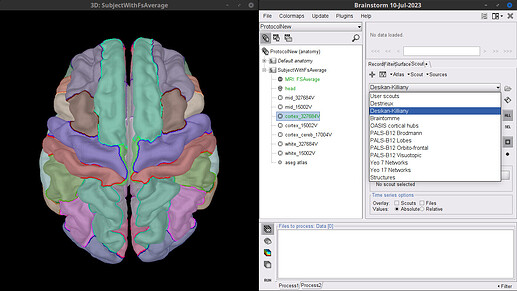
If the w-files were computed with the FsAverage. You can create a new Subject in Brainstorm, and use the FsAverage template (right-click on Subject > Use template > FsAverage > FsAverage_2020). In the template files, the surface file cortex_327684 has already cortical parcellations:
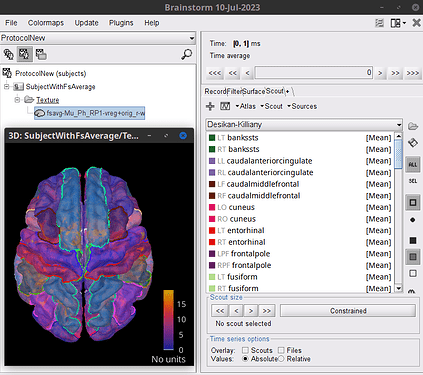
Indeed, you would need to import the w-files as textures (to the Subject using the FsAverage template) then it will be possible to explore the w-file content per Scout (ROI). The index of the vertices for each Scout are found in the Surface file as @tmedani showed: About vertices in surface file - #3 by tmedani
It is needed to also add the times for each column in the Time field. E.g., [0,1,2]