Hi,
I am trying to import the anatomy freesurf folders by 'brainstorm3.command /opt/mcr/v91 **.m /input/MRI/04' on my Linux system. But after the procedure, in the anat directory, there are only three files(brainstormsubject.mat, subjectimage_t1.mat, tess_1h_pial.mat), and no errors were shown. I also run the script in matlab, everything is fine.
So is there anyone who knows the reasons?
What is "**.m"?
Is it a script you wrote?
Can you post here it contains?
Actually, it's readin_mri.m written by me.
when I run it in Linux, the line 'brainstorm nogui' was commented.
The matlab version is R2016b and the mcr version is v91.
In matlab, I run it by: readin_mri('G:\MEG_process\algorithm\meg_pipeline\input\MRI\04','04').
In Linux, I run it by:
/opt/brainstorm3/bin/R2016b/brainstorm3.command /opt/mcr/v91 readin_mri.m /input/MRI/04 04
Here is the script:
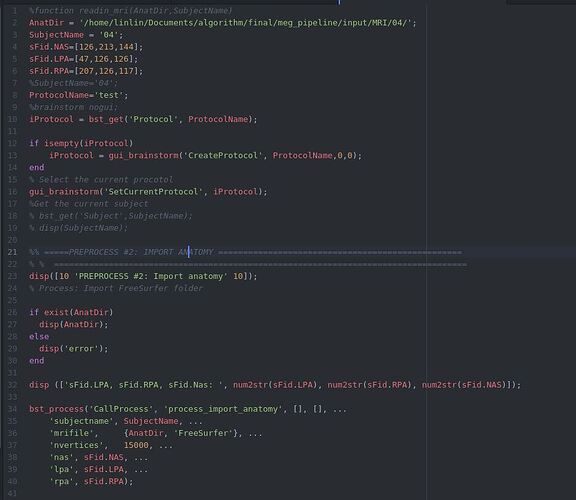
function readin_mri(AnatDir,SubjectName)
sFid.NAS=[126,213,144];
sFid.LPA=[47,126,126];
sFid.RPA=[207,126,117];
%SubjectName='04';
ProtocolName='test';
%brainstorm nogui;
iProtocol = bst_get('Protocol', ProtocolName);
if isempty(iProtocol)
iProtocol = gui_brainstorm('CreateProtocol', ProtocolName,0,0);
end
% Select the current procotol
gui_brainstorm('SetCurrentProtocol', iProtocol);
%Get the current subject
bst_get('Subject',SubjectName);
disp(SubjectName);
%% =====PREPROCESS #2: IMPORT ANATOMY =================================================
% % ===================================================================================
disp([10 'PREPROCESS #2: Import anatomy' 10]);
% Process: Import FreeSurfer folder
disp (['sFid.LPA, sFid.RPA, sFid.Nas: ', num2str(sFid.LPA), num2str(sFid.RPA), num2str(sFid.NAS)]);
bst_process('CallProcess', 'process_import_anatomy', [], [], ...
'subjectname', SubjectName, ...
'mrifile', {AnatDir, 'FreeSurfer'}, ...
'nvertices', 15000, ...
'nas', sFid.NAS, ...
'lpa', sFid.LPA, ...
'rpa', sFid.RPA);
I've tested your script with the compiled version of Brainstorm and realized there were maybe some issues with different issues of the database. I recompiled brainstorm for Matlab 2015b (the default version) and 2016b and updated them on the website.
I could use your script to import the FreeSurfer anatomy for the example dataset of the introduction tutorials. It works fine and imports everything.
First try updating Brainstorm both on Windows (script version: use the menu Update > Update Brainstorm from the Brainstorm interface) and on Linux (compiled version: delete the brainstorm3 folder and download a new one).
If you still have the same problem, it could be:
- Because of this specific FreeSurfer folder: do you have the same issue with all your subjects?
- Because there are compatibility issues between the two environments. Is this the same shared network folder you are using in both environments? I've experienced weird issues in this configuration previous, try creating a new protocol in your script in the compile Linux version (instead of reusing one created on Windows), and see whether it helps.
FYI: the line "bst_get('Subject',SubjectName);" in your script has no effect.
Thank you for the detailed information.
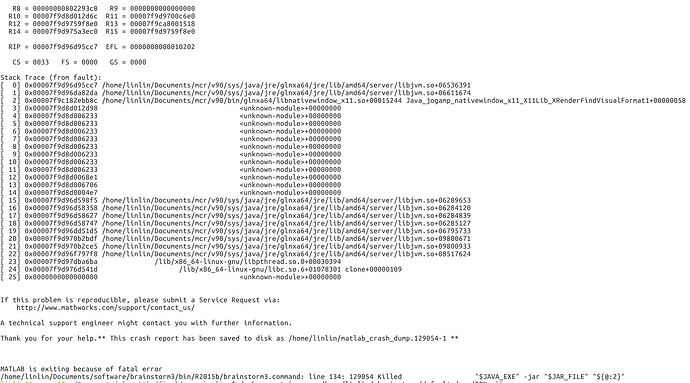
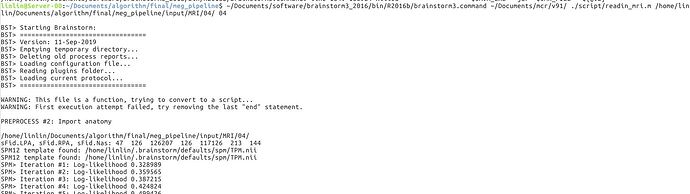
I tested the script after update brainstorm on Linux system, and I still cannot run it correctly. The error is as follows:
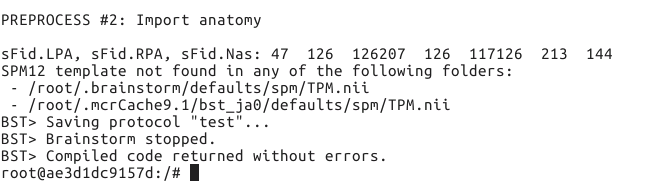
OK, so the error is due to the SPM atlas TPM.nii not being found in the compiled code.
Is this really the version I compiled yesterday?
Can you try to locate TPM.nii in /root/.mcrCache9.1
Please post all the text output from the console, starting from the command you executed.
Can you please try with the 2015b version? (the one packaged with Brainstorm - you need to install the matching MCR)
Maybe there is an issue with the way the path are managed in 2016b, but I've just tested it on windows and it seems to work fine.
Btw, you should not be running this as root. Maybe there are issues related with this as well.
Try reinstalling everything as a regular user.
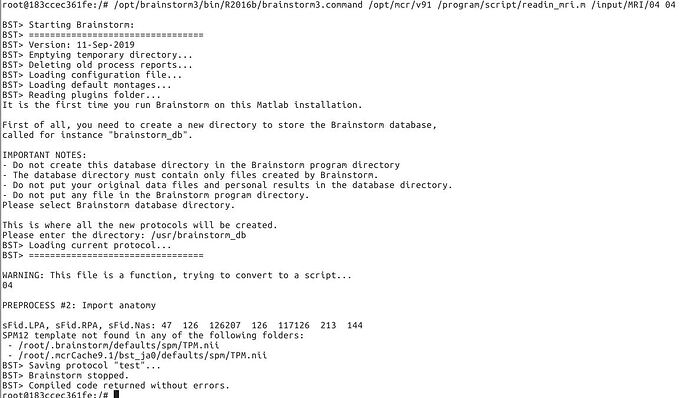
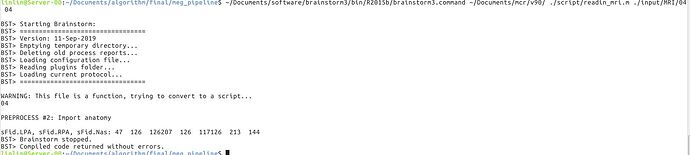
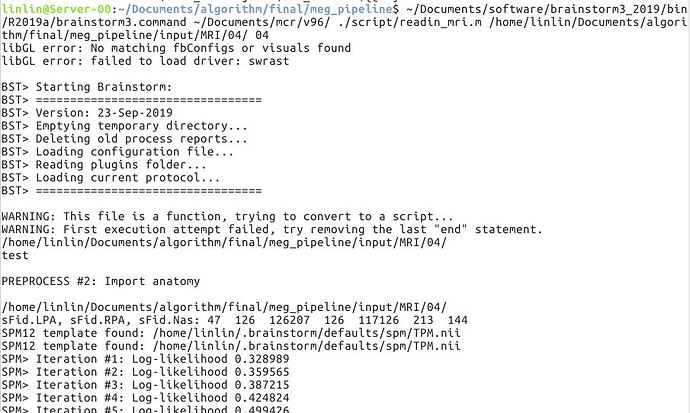
The first figure is the error with 2016b and root. The mcr and brainstorm are reinstalled.
The second one is that with 2015b and other user. After importing, there is only one file in the anat directory. That's brainstormsubject.mat.
And I also try this on another subject data, it cannot work either.
"This file is a function, trying to convert to a script"
=> Remove the function header in your .m file
Have you tried loading this file from the interface instead? (still using the compiled version)
If the function header is removed, how can I transfer the parameters to the script?
What's your meaning of 'loading this file from the interface (still using the compiled version)'.
I imported the folder from brainstorm interface, everything was fine.
I also tested the script by matlab, which is: run it in matlab command environment by 'readin_mri('/input/MRI/04', '04')' with 'brainstorm nogui' uncommented. All the files can be imported correctly.
You can't execute functions when you don't have a Matlab license. A program compiled with the Matlab compiler is not able to execute any code that was not compiled together with the program. It makes sense in terms of licensing, from the company's point of view...
The way we execute Brainstorm scripts from the compiled distribution does not rely on the Matlab's regular execution of scripts/functions (what is used when you call your script from the Matlab command window), but we read the .m file and execute it with the eval() function. This is a solution that offers a lot less possibilities, but it's still better than nothing...
This is documented in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Running_scripts_on_a_cluster
Try rewriting it as a script with the parameters at the beginning, with as few lines as possible, and try to understand where the problem is coming from.
Thanks for the suggestion.
I wrote the parameters into the script. And the error still showed.
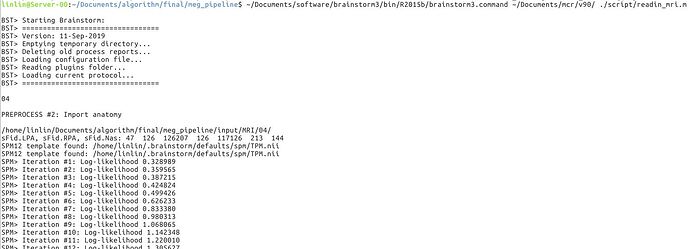
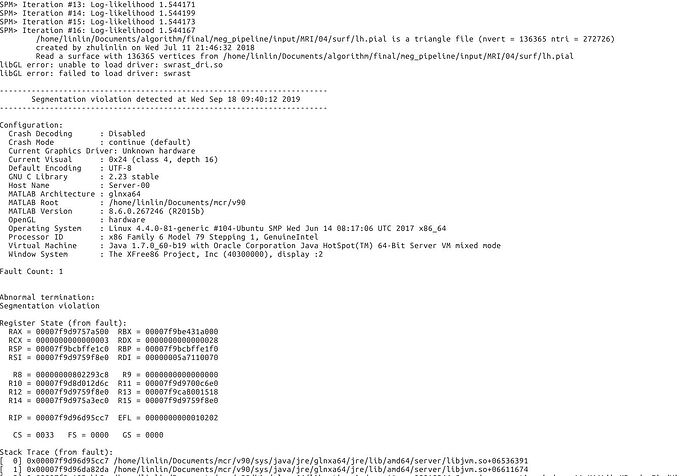
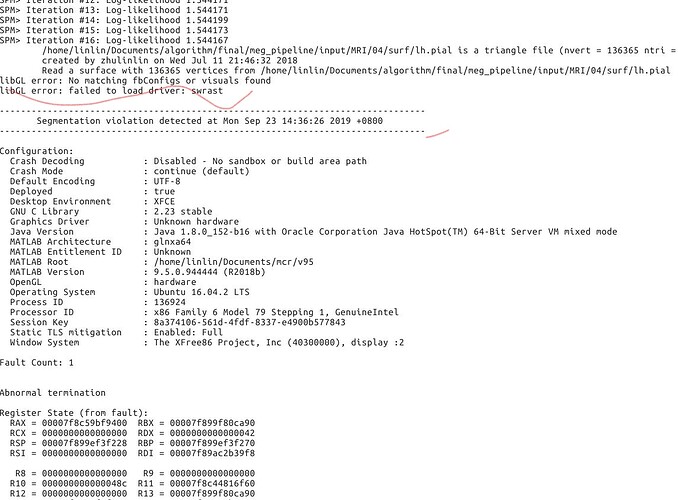
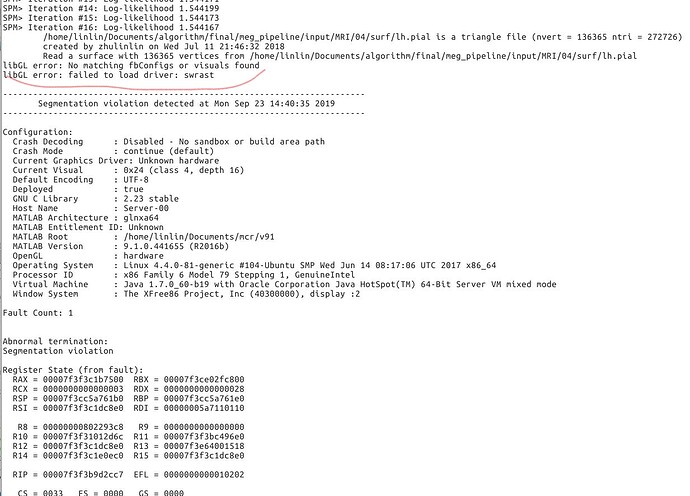
So I copied a TPM.nii from spm12 to the path. The error is shown as followed. Is there any more suggestion? This seems like a hardware error?
It crashes probably because it can't access the display or initialize any OpenGL driver (libGL error).
Start Brainstorm with "brainstorm server" instead of "brainstorm nogui" for avoiding any access to the display:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Running_scripts_on_a_cluster
I do run the script by 'brainstorm.command readin_mri.m', from the tutorials, it's a server mode.
This is the script.
Indeed, this is already running in server mode.
I'm not sure I understand what requires the loading of the OpenGL driver then...
Can you try with newer versions of Matlab?
I recompiled Brainstorm for 2018b and 2019a so you can test them, they are available on the download page.
(you need to download the MCR for each of them, sorry for the complicated debugging procedure...)
Hi, Francois,
Thank you for the patient.
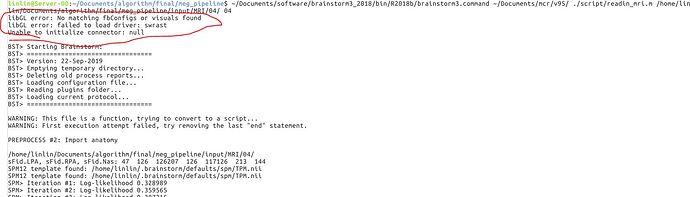
First of all, I tested the script with 2018b and 2019a, the errors are as same as the previous version. As shown in the following figure.
And then, I think maybe it's because there are some problems with the lib. I searched the forum of matlab, and found this: https://ww2.mathworks.cn/matlabcentral/answers/241374-libgl-error-persists-and-i-ve-tried-everything
But, I modified the lib in Linux system as the suggestions. The errors are still there. (it seems like a little bit different with the previous ones Before I modified these libs, the errors had been the same with those in matlab 2018b and 2019a). I checked my server and found there are no graphic card and graphic driver. So, Is the importing procedure involved with some GUI processing?
Your system cannot load the "swrast" library, which is responsible for the software OpenGL rendering. But even without a display or a graphics card, the software OpenGL should be usable.
Therefore there is definitely a problem on your system: coming either from Matlab, or from the libraries installed on your system, or from the incompatibilities between the two.
Posting the full crash report as a bug report on the Mathworks website may help us understand what is the source of the problem on your system. Maybe installing/updating the MESA driver on your system could solve the problem (there are lots of forum posts on this topic all over the internet).
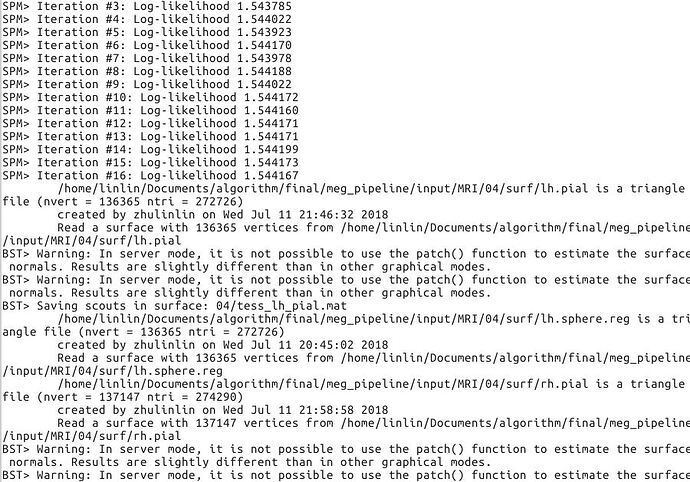
But this wouldn't solve the Brainstorm part: why does Matlab need to load the OpenGL drivers immediately after importing the surface? I may have found the cause of the problem in the way the surface normals are computed (tess_normals): it was using the patch() function to create a surface and compute the normals, which might be done by the software OpenGL driver... I replaced this piece of code with a different estimation of the normals, when in server mode. The results are a bit different, therefore the source estimation would be slightly different than in graphics mode, but at least it might work.
Here is corresponding GIT commit:
https://github.com/brainstorm-tools/brainstorm3/commit/c4cbfb2cca3ed533df2f29709219f3005ff48c46
I recompiled the 2015b and 2019a versions with this modification.
Try first with the standard version (2015b, download the default Brainstorm package), and if it doesn't work try with the 2019a.
Hi Francois,
Thank you for the modification. Both versions can work on the Linux system while at the very first processing of 2019 version the same errors are shown (swrast error). But it seems no effect on the following computing.
Could you compile a 2016b version? Because I have much more script based on the 2016b version.
Done, today's 2016b binaries are online as well.
Please let us know how this goes.
Thank you very much. It works.