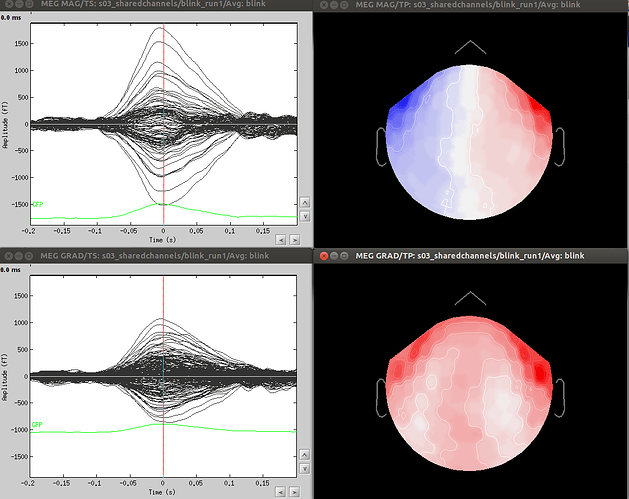
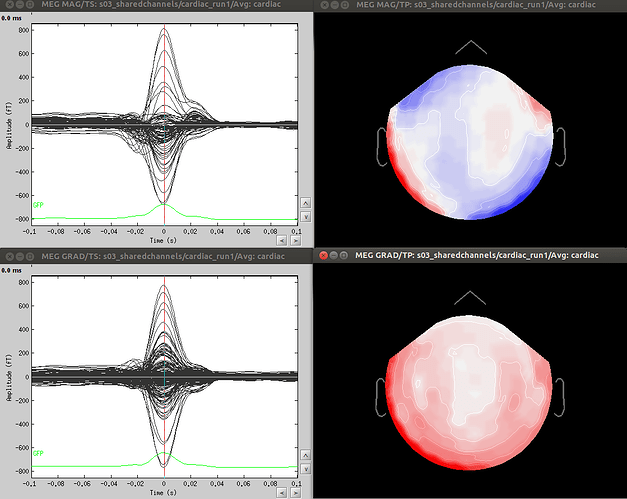
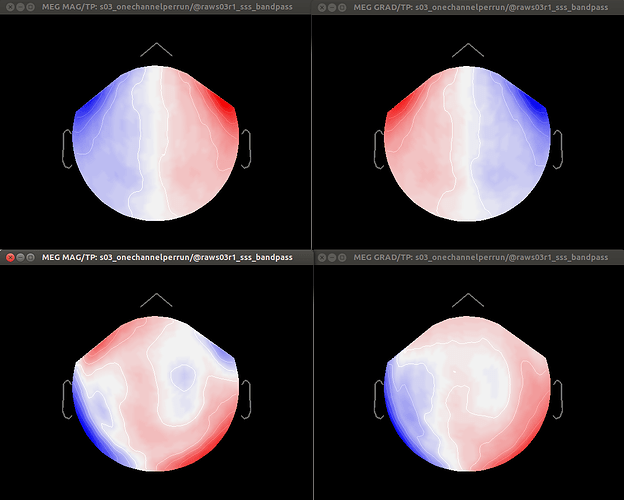
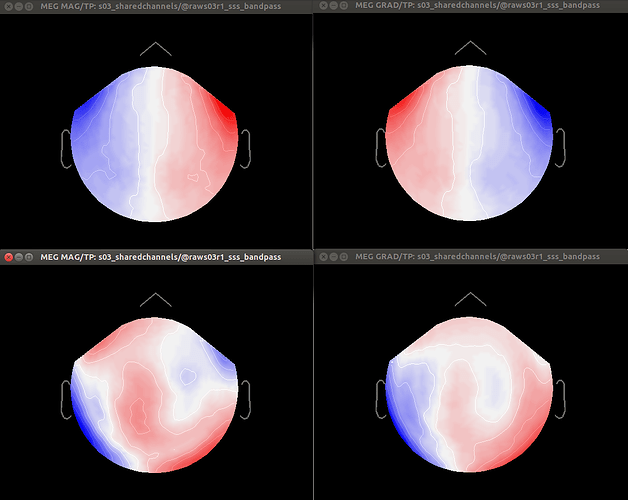
Dear Dimitrios and all, in particular Neuromag users,
I am still terribly confused as to what is the best strategy to use to compute SSP projectors for blink/cardiac artifact removal in Neuromag data. Typical datasets from my lab contain 4 to 8 separate runs from each subjects.
One source of complication is that for Neuromag data, co-registration of head positions across several runs is usually computed at the first preprocessing step together with Maxfilter, and blink/ECG SSPs are usually computed afterwards.
I wonder whether:
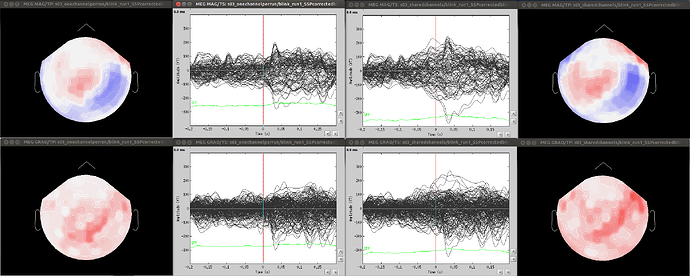
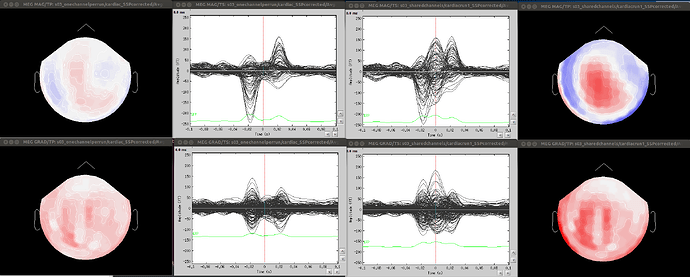
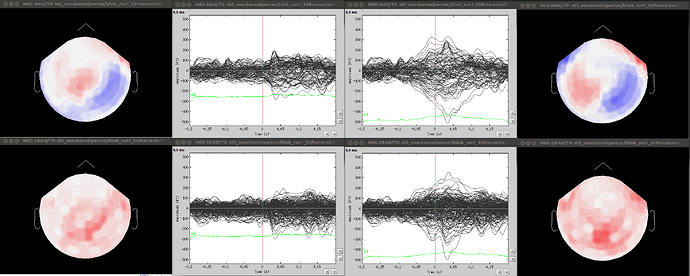
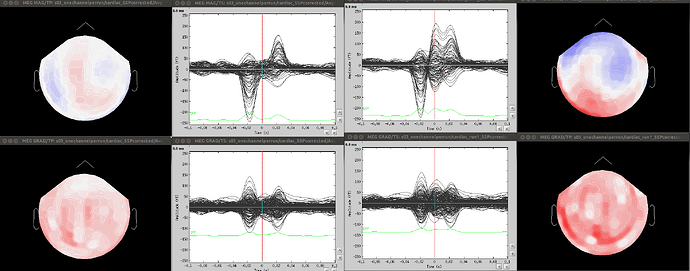
- Since co-registration has already been computed, does it make sense to compute SSPs for each run separately, or it’s reliable to compute it on all runs together? As Francois suggested in a previous thread, this could be done by extracting blink epochs from all runs and compute the SSP on that:
http://neuroimage.usc.edu/forums/showthread.php?1113-EOG-ECG-SSP-over-several-runs&p=4810#post4810
Also, this would allow (contrary to the first solution) using one channel set per subject, which is much more practical for the following steps.
- Otherwise, should I first apply Maxfilter WITHOUT Maxmove, compute SSPs over each run, and only then co-register by applying Maxmove? And what would happen to SSP projections in this case?
Thanks for your feedback,
Marco
[QUOTE=pantazis;5661]Hi Francois,
I understand this is a very complicated issue.
Brainstorm stores the projectors inside the channel file because they are used for forward modeling. I would like to argue that this is not the proper way, because it ties subject registration to projectors, which are two very different processes.
-projectors are created using noise covariance (ssp), eyeblinks (svd on data templates), heartbeats, etc. In all the cases that come to mind (and correct me if I am wrong), projectors are defined from the MEG data without any dependence on the subject registration.
-Subject registration changes the position of the channels, but does not affect the projectors.
By combining the two in the same file, there are problems. On multiple occasions, (refining subject registration using polhemus points etc), I want to revert to the previous registration (nice if such option existed), but the only way to do so is to reimport the channel file. But by importing a channel file (directly from the fif file), I lose all my projectors. Since misregistration is worse than suboptimal modeling, I always opt to correct registration.
To resolve this issue, I recommend saving the projectors in a separate file, always next to a channel file.
Regarding the suggestion ‘you should have one channel file per run in your database, and you should calculate your projectors individually for each FIF file’. It is not that simple. Nearly all our MEG scans result in several fif files that:
-share the same registration. (A) several short recordings for the same subject produce different fif files that are aligned using the maxfilter trans option; or (B) a single long run produces multiple fif files because the acquisition computer cannot handle large files)
-share the same projectors (eyeblinks, ssp etc).
Requiring separate channels and projectors per fif file is not practical, because most of the times they share this information.
Minor comment: Also, calculating projectors separately per fif file can produce unstable results (case A).
I guess all the above problems would be solved if there was a way to combine several fif files together. Then there would be a single fif file per run (rather than several with the same registration), and no changes in Brainstorm would be necessary. Do you know a software that combines different fif files into one?
Thank you,
Dimitrios
Best,
Dimitrios[/QUOTE]