I am unclear how to proceed with the channel files though. When trying to add them, I got an error
You can ignore this warning. Brainstorm expects to find recordings in FIF files. There are none, it complains, but it does not cause any additional problem.
I think only one channel set can be loaded in a subject at a given time, right?
No. You should create one subfolder for each template (and create one channel file per folder).
Also, you should start from an existing template, not from an empty structure.
The order of the digitized points in the .fif file should match the electrodes in the Brainstorm template (no other solution, since your 3D points are not named in the fif file...). Or we need to add some code to re-order them.
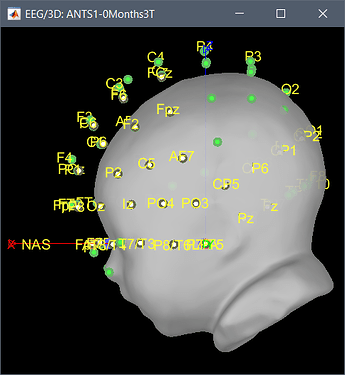
It seems that there are issues in the electrodes positions you provide (I tested only ANTS1-0Months3T):
- The order of the electrodes is incorrect
- The NAS/LPA/RPA does not seem correct (but I don't have your fiducials.mat so I can't test this part properly)
- The size of the cap does not match the size of the head.
Below: your script modified to create one channel file per folder.
% Get EEG defaults folder
RefChannelPath = bst_fullfile(bst_get('BrainstormDefaultsDir'), 'eeg', 'ICBM152');
files = ...; % Keep your original code
fid_dict = load('/home/christian/brainstorm_db/fiducials.mat');
delete '/home/christian/.brainstorm/defaults/anatomy/*'
for iSubj = 1:size(files, 1)
% Create subject
SubjectName = files(iSubj).name;
[sSubject, iSubject] = db_add_subject(SubjectName, [], 0, 0);
% Import anatomy
FsDir = bst_fullfile(files(iSubj).folder, files(iSubj).name);
sFid = ...; % Keep your original code
import_anatomy_fs(iSubject, FsDir, 15000, 0, sFid);
% Create folder for channel file
for iMontage = 1:5
switch iMontage
case 1
MontageName = '10-5';
RefChannelFile = 'channel_ASA_10-05_343.mat';
case 2
MontageName = '10-10';
RefChannelFile = 'channel_10-10_65.mat';
case 3
MontageName = '10-20';
RefChannelFile = 'channel_10-20_19.mat';
case 4
MontageName = 'HGSN128';
RefChannelFile = 'channel_GSN_HydroCel_128_E1.mat';
case 5
MontageName = 'HGSN128';
RefChannelFile = 'channel_GSN_HydroCel_128_E001.mat';
end
% Load reference channel file
RefChannelMat = in_bst_channel(bst_fullfile(RefChannelPath, RefChannelFile));
FolderName = file_standardize(str_remove_parenth(RefChannelMat.Comment));
% Create new folder
iStudy = db_add_condition(SubjectName, FolderName);
sStudy = bst_get('Study', iStudy);
% Read channel file
FifMat = import_channel([], bst_fullfile(FsDir, [MontageName '-montage.fif']), 'FIF', 0, 0, 0);
% Create destination channel file
ChannelMat = RefChannelMat;
ChannelMat.SCS.LAS = FifMat.HeadPoints.Loc(:,1);
ChannelMat.SCS.NAS = FifMat.HeadPoints.Loc(:,2);
ChannelMat.SCS.RPA = FifMat.HeadPoints.Loc(:,3);
for i = 15:size(FifMat.HeadPoints.Loc, 2)
% TODO: Fix order of electrodes
if (i-14 <= length(ChannelMat.Channel))
ChannelMat.Channel(i-14).Loc = FifMat.HeadPoints.Loc(:,i);
end
end
ChannelMat.HeadPoints = FifMat.HeadPoints;
% Re-Register based on NAS/LPA/RPA
% ChannelMat = channel_detect_type(ChannelMat, 1, 0);
% Save channel file
ChannelFile = bst_fullfile(bst_fileparts(file_fullpath(sStudy.FileName)), RefChannelFile);
bst_save(ChannelFile, ChannelMat, 'v7');
db_reload_studies(iStudy);
end
bst_memory('UnloadAll', 'Forced');
db_reload_subjects(iSubject);
panel_protocols('UpdateTree');
db_save();
export_default_anat(iSubject, subjectName, 1);
end