Hi Christian,
Just a quick check. Should I email John Richard to see if he will be OK to share his template (processed by you). I do want to test the templates in brainstorm soon. Please let me know what you think.
Thanks!
Yuhan
Hi Christian,
Just a quick check. Should I email John Richard to see if he will be OK to share his template (processed by you). I do want to test the templates in brainstorm soon. Please let me know what you think.
Thanks!
Yuhan
@yuhanmc No, it is fine with John. I am in touch with him. I can send you a version of it now if you'd like, but be advised that it might not be the last version yet. We are still figuring out some little details. I think that were are still somewhat like 1-2 weeks from a final version. It should already be not bad thought, and if your pipeline is mostly automated, you should be able to get going with these and just swap the final version when it is ready. Do you want to proceed that way? If so, I'll send you this preliminary version directly by email.
@christianoreilly That would be great and I will swap the final version later. I am in the middle of comparing source reconstruction results using different age template. So I certainly would love to try your current version even if it is not final. I would try it on a few subjects at different age and when your template is final, I will use them on all the rest of the infant MEG data.
So yes please email this version to me and I can also test it and let you know what comes out.
Thank you for willing to share this !!
Best, Yuhan
Hi Christian! What a wonderful contribution! Thank you! We are currently analyzing data from infants aged 3 to 6 months. We are using Brainstorm/Nirstorm since we are analyzing fNIRS data, and we would like to use the templates you created. If you are willing to share the templates, you can send a link to my email (sophie.l.bouton@gmail.com). Thank you !
Sophie
Just a small update for you @Francois and to whomever if following this post: I have finished preparing 12 templates from 0 to 2 years old. I wrote an initial draft of a paper presenting them. I would consider these templates as being at the "beta" release stage. If anyone is interested in testing/using them, they can contact me directly. Maybe we can add them officially to Brainstorm when we get closer to the publication of this paper? That would give chance to beta-test it a bit more. Does that sounds reasonable @Francois ?
That would be fantastic, Christian!
Yes, that would be awesome!
There is a menu to package an anatomy folder as a template: it removes all unnecessary fields from the files (like the interpolation matrices, the VertConn field in the surfaces, the history) and creates a .zip file.
In the anatomy view, right-click on the subject > Use template > Create new template.
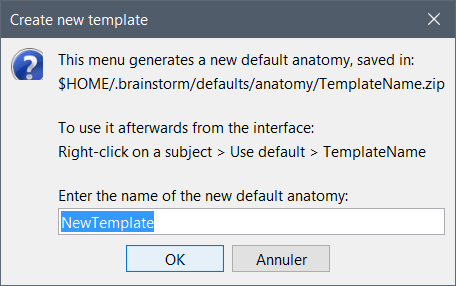
When you get the 12 .zip files ready, please upload them somewhere, share the download link here, and I'll upload them on the Brainstorm server and make them available for download for all the Brainstorm users.
We will also need to add some description of the templates, including the reference to your article, on this page:
https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy
Thanks!
Yes, I saw this feature. I used it to "package" the 12 templates for Brainstorm. Very convenient. I'll provide some text that describes the templates for the tutorial page and email you a link to retrieve the zip files as soon we get closer to publication.
@christianoreilly Thank you for the contribution. Just want to let you know that I have been using the templates you sent to me. I will keep you updated as I tested them out on different protocols and different analyses pipelines (MNE, or PSD in source space). The best thing about it is that they all shared the same number of vertices across age with the same atlases. So it is really a great feature to to look at source level data in a longitudinal/cross-sectional cohorts from 0 - 2 years old!
Best, Yuhan
Geat! Happy that it is useful for your work @yuhanmc Let me know if you meet issues with them.
@christianoreilly Should I wait before downloading the new templates?
Sorry for the false alarm! I hoped it would pass under the radar  Yeah, hold on just a few days please. Everything was done and ready but then someone noticed and error for one of the template so I will just reprocess this template and re-upload. I'll re-post the message when it is done.
Yeah, hold on just a few days please. Everything was done and ready but then someone noticed and error for one of the template so I will just reprocess this template and re-upload. I'll re-post the message when it is done.
[I deleted this message previously because I needed to correct an issue. Reposting now because the issue is now solved and we are good to go. ]
@Francois I think it is all ready to go now. The new template can be found here: https://drive.google.com/file/d/1qpEMUaE1Lz_aPOxKgcwi4MHWEZXhkds_/view?usp=sharing
These are 13 templates from 2 weeks to 2 years of age. The following paper (OA preprint) describes them in detail: https://www.biorxiv.org/content/10.1101/2020.06.20.162131v1 (@yuhanmc @SophieBouton If you need more info on the templates I previously shared with you, you will find it in this paper. )
Let me know when it is merged in Brainstorm so that I can remove the archive from my GDrive. Once these have been added, I would suggest removing the previous 1-yo template that I provided since I have full confidence that these new templates are of higher quality and should therefore be used instead.
To the best of my knowledge, all is good with these templates. But please let me know if you experience any issues.
Hi Christian, and All,
Can you please tell, how can we load the template in brainstorm (step by step)
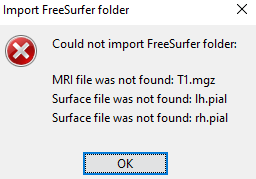
If I make import anatomy folder I have this error:
If I make import MRI I can only have the subjectimage_T1:
so I am not succeed to import all of these:
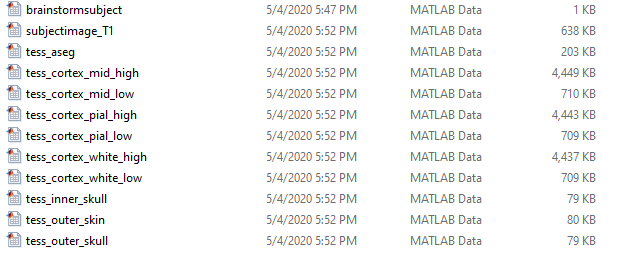
Thank you for help
Thank you so much, Christian!
François is away for the week. As he has worked with you closely on this, let's wait until he is back to finalize the integration in the distribution, if that's ok from your end.
Sure @Sylvain, there is nothing pressing for me.
@abdallahqusaibe This is not done by loading the anatomy folder.
First unzip the main archive. It will contain 13 zip archives. Place these 13 archives in $home/.brainstorm/default/anatomy/ Then, to use these template, right click the default anatomy specification in your project and in the contextual menu select "Use template" and chose whichever template you want to use from this list. Else, if there is no emergency on your side for these analyses, you can wait for Francois to integrate it directly to the Brainstorm distribution.
Done: https://github.com/brainstorm-tools/brainstorm3/commit/477a07437bc5ec25489ed53d2c5687d6c392e0be
Update Brainstorm to access these new templates.
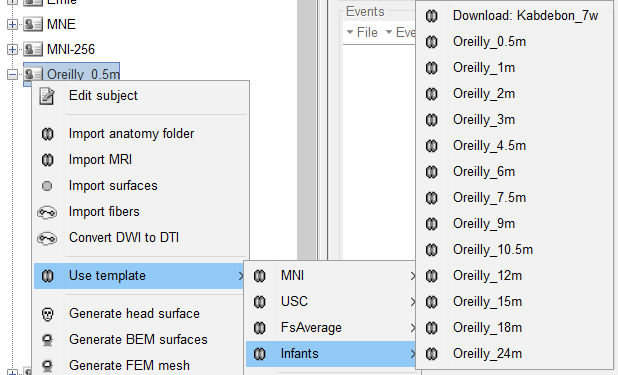
I updates the documentation, please let me know if you'd like to modify something:
https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#FreeSurfer_templates
There are apparently issues with the ASEG atlas for 0.5m, 1m, 2m and 3m:
Are you fixing this, or should we just delete these ASEG files for these templates?
@christianoreilly
Do you happen to have electrodes positions available for all these babies? 10-10 or EGI?
This would save a lot of time for people trying to use them for source analysis.
@christianoreilly
Do you happen to have electrodes positions available for all these babies?
Hi @francois, sorry for my unresponsiveness. I initially computed these for Brainstorm but the saving as atlas was not packaging the computed electrode positions with the surfaces and volumes saved in the template so I stopped putting time into computing them. I have electrode positions computed with MNE as fif objects. Are these importable in Brainstorm? I can send you these fif montage if you want. Else, I would need to convert them in a brainstorm acceptable format and ensured that the coregister with the surfaces was preserved by the process of importation.